Chlamydiais agenusofpathogenicGram-negative bacteriathat areobligate intracellular parasites.Chlamydiainfectionsare the most common bacterialsexually transmitted diseasesin humans and are the leading cause of infectious blindness worldwide.[2]
| Chlamydia | |
|---|---|
| |
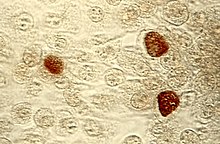
| Chlamydia trachomatisinclusion bodies(brown) in a McCoy cell culture. | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Chlamydiota |
| Class: | Chlamydiia |
| Order: | Chlamydiales |
| Family: | Chlamydiaceae |
| Genus: | Chlamydia Jones, Rake & Stearns 1945 |
| Type species | |
| Chlamydia trachomatis (Busacca 1935) Rake 1957
| |
| Species[1] | |
| Synonyms | |
| |
SpeciesincludeChlamydia trachomatis(a human pathogen),Ch. suis(affects onlyswine), andCh. muridarum(affects onlymiceandhamsters).[3]Humans mainly contractCh. trachomatis,Ch. pneumoniae,Ch. abortus,andCh. psittaci.[4]
Classification
editBecause ofChlamydia's unique developmental cycle, it was taxonomically classified in a separate order.[5]Chlamydiais part of the order Chlamydiales, family Chlamydiaceae.[citation needed]
In the early 1990s six species ofChlamydiawere known. A major re-description of the Chlamydiales order in 1999, using the then new techniques of DNA analysis, split three of the species from the genusChlamydiaand reclassified them in the then newly created genusChlamydophila,and also added three new species to this genus.[6]In 2001 many bacteriologists strongly objected to the reclassification,[1]although in 2006 some scientists still supported the distinctness ofChlamydophila.[7]In 2009 the validity ofChlamydophilawas challenged by newer DNA analysis techniques, leading to a proposal to "reunite theChlamydiaceaeinto a single genus,Chlamydia".[8]This appears to have been accepted by the community,[9][10]bringing the number of (valid)Chlamydiaspecies up to 9. Many probable species were subsequently isolated, but no one bothered to name them. In 2013 a 10th species was added,Ch. ibidis,known only from feralsacred ibisin France.[11]Two more species were added in 2014 (but validated 2015):Ch. aviumwhich infects pigeons and parrots, andCh. gallinaceainfecting chickens, guinea fowl and turkeys.[4]Ch. abortuswas added in 2015, and theChlamydophilaspecies reclassified.[1]A number of new species were originally classified as aberrant strains ofCh. psittaci[4]
Genomes
editChlamydiaspecies have genomes around 1.0 to 1.3megabasesin length.[12]Most encode ~900 to 1050 proteins.[13]Some species also contain a DNAplasmidsorphagegenomes (see Table). The elementary body contains an RNA polymerase responsible for the transcription of the DNA genome after entry into the host cell cytoplasm and the initiation of the growth cycle.Ribosomesand ribosomal subunits are found in these bodies.[14]
| Ch. trachomatisMoPn | Ch. trachomatisD | Ch. pneumoniaeAR39 | Ch. pneumoniaeCWL029 | |
|---|---|---|---|---|
| Size (nt) | 1,069,412 | 1,042,519 | 1,229,853 | 1,230,230 |
| ORFs | 924 | 894 | 1052 | 1052 |
| tRNAs | 37 | 37 | 38 | 38 |
| plasmids | 1 (7,501 nt) | 1 (7,493 nt) | 1 ssDNA phage | none |
Table 1.Genome features of selectedChlamydiaspecies and strains. MoPn is a mouse pathogen while strain "D" is a human pathogen. About 80% of the genes inCh. trachomatisandCh. pneumoniaeare orthologs. Adapted after Read et al. 2000[13]
Developmental cycle
editChlamydiamay be found in the form of an elementary body and a reticulate body. The elementary body is the nonreplicating infectious particle that is released when infected cells rupture. It is responsible for the bacteria's ability to spread from person to person and is analogous to aspore.The elementary body may be 0.25 to 0.30 μm in diameter. This form is covered by a rigidcell wall(hence thecombining formchlamyd-in the genus name). The elementary body induces its ownendocytosisupon exposure to target cells. Onephagolysosomeusually produces an estimated 100–1000 elementary bodies.[citation needed]
Chlamydiamay also take the form of a reticulate body, which is in fact anintracytoplasmicform, highly involved in the process of replication and growth of these bacteria. The reticulate body is slightly larger than the elementary body and may reach up to 0.6 μm in diameter with a minimum of 0.5 μm. It does not have a cell wall. When stained withiodine,reticulate bodies appear as inclusions in the cell. The DNA genome, proteins, and ribosomes are retained in the reticulate body. This occurs as a result of the development cycle of the bacteria. The reticular body is basically the structure in which the chlamydial genome is transcribed into RNA,proteinsare synthesized, and the DNA is replicated. The reticulate body divides by binary fission to form particles which, after synthesis of the outer cell wall, develop into new infectious elementary body progeny. The fusion lasts about three hours and the incubation period may be up to 21 days. After division, the reticulate body transforms back to the elementary form and is released by the cell byexocytosis.[5]
Studies on the growth cycle ofCh. trachomatisandCh. psittaciincell culturesin vitroreveal that the infectious elementary body (EB) develops into a noninfectious reticulate body (RB) within a cytoplasmic vacuole in the infected cell. After the elementary body enters the infected cell, an eclipse phase of 20 hours occurs while the infectious particle develops into a reticulate body. The yield of chlamydial elementary bodies is maximal 36 to 50 hours after infection.[14]
A histone like protein HctA and HctB play role in controlling the differentiation between the two cell types. The expression of HctA is tightly regulated and repressed by small non-coding RNA, IhtA until the late RB to EB re-differentiation.[15]The IhtA RNA is conserved acrossChlamydiaspecies.[16]
Pathology
editMost chlamydial infections do not cause symptoms.[17]Symptomatic infections often include a burning sensation when urinating and abdominal or genital pain and discomfort.[18]All people who have engaged in sexual activity with potentially infected individuals may be offered one of several tests to diagnose the condition.[citation needed]Nucleic acid amplification tests(NAAT), which includepolymerase chain reaction(PCR),transcription-mediated amplification(TMA),ligase chain reaction(LCR), andstrand displacement amplification(SDA), are the most widely used diagnostic test forChlamydia.[19]
Evolution
editRecentphylogeneticstudies have revealed thatChlamydialikely shares acommon ancestorwithcyanobacteria,the group containing theendosymbiontancestor to thechloroplastsof modernplants,hence,Chlamydiaretains unusual plant-like traits, bothgeneticallyandphysiologically.In particular, theenzymeL,L-diaminopimelate aminotransferase,which is related tolysineproduction in plants, is also linked with the construction of chlamydialpeptidoglycan,which is required for division.[20]The genetic encoding for the enzymes is remarkably similar in plants, cyanobacteria, andChlamydia,demonstrating a close common ancestry.[21]
Phylogeny
edit| 16S rRNA basedLTP_08_2023[22][23][24] | 120 marker proteins basedGTDB08-RS214[25][26][27] | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
See also
editReferences
edit- ^abcParte, A.C."Chlamydia".LPSN.
- ^Drew, W. Lawrence (2004). "Chlamydia". In Ryan, Kenneth; Ray, C. George (eds.).Sherris Medical Microbiology(PDF)(4th ed.). McGraw Hill. pp. 463–470.ISBN978-0-8385-8529-0.
- ^Ward M."Taxonomy diagram".Chlamydiae.com.Archived fromthe originalon 2010-09-18.Retrieved2008-10-28.
- ^abcJoseph, SJ; et al. (2015), "Chlamydiaceae genomics reveals interspecies admixture and the recent evolution of Chlamydia abortus infecting lower mammalian species and humans",Genome Biol Evol,7(11): 3070–3084,doi:10.1093/gbe/evv201,PMC4994753,PMID26507799.
- ^ab"Chlamydia trachomatis".Archived fromthe originalon July 2, 2010.RetrievedJune 18,2010.
- ^Everett KD, Bush RM, Andersen AA (April 1999)."Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms".Int. J. Syst. Bacteriol.49(2): 415–40.doi:10.1099/00207713-49-2-415.PMID10319462.
- ^Griffiths E, Ventresca MS, Gupta RS (2006)."BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species".BMC Genomics.7:14.doi:10.1186/1471-2164-7-14.PMC1403754.PMID16436211.
- ^Stephens RS, Myers G, Eppinger M, Bavoil PM (March 2009)."Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved".FEMS Immunol. Med. Microbiol.55(2): 115–9.doi:10.1111/j.1574-695X.2008.00516.x.PMID19281563.
- ^Greub, Gilbert (1 November 2010)."International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of the Chlamydiae Minutes of the inaugural closed meeting, 21 March 2009, Little Rock, AR, USA".International Journal of Systematic and Evolutionary Microbiology.60(11): 2691–2693.doi:10.1099/ijs.0.028225-0.PMID21048221.
- ^Balsamo, G; et al. (2017),"Compendium of measures to control Chlamydia psittaci infection among humans (psittacosis) and pet birds (avian chlamydiosis), 2017"(PDF),J Avian Med Surg,31(3): 262–282,doi:10.1647/217-265,PMID28891690,S2CID26000244.
- ^Vorimore, Fabien; Hsia, Ru-ching; Huot-Creasy, Heather; Bastian, Suzanne; Deruyter, Lucie; Passet, Anne; Sachse, Konrad; Bavoil, Patrik; Myers, Garry; Laroucau, Karine (20 September 2013)."Isolation of a NewChlamydiaspecies from the Feral Sacred Ibis (Threskiornis aethiopicus)-Chlamydia ibidis".PLOS ONE.8(9). e74823.Bibcode:2013PLoSO...874823V.doi:10.1371/journal.pone.0074823.PMC3779242.PMID24073223.
- ^"EMBL bacterial genomes".RetrievedJanuary 19,2012.
- ^abRead, T. D.; Brunham, R. C.; Shen, C.; Gill, S. R.; Heidelberg, J. F.; White, O.; Hickey, E. K.; Peterson, J.; Utterback, T. (2000-03-15)."Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39".Nucleic Acids Research.28(6): 1397–1406.doi:10.1093/nar/28.6.1397.ISSN1362-4962.PMC111046.PMID10684935.
- ^abBecker, Yechiel (1996)."Chlamydia".In Baron, S. (ed.).Medical Microbiology(4th ed.). University of Texas Medical Branch at Galveston.ISBN0-9631172-1-1.PMID21413294.
- ^Grieshaber, NA; Grieshaber, SS; Fisher, ER; Hackstadt, T (2006). "A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis".Mol. Microbiol.59(2): 541–50.doi:10.1111/j.1365-2958.2005.04949.x.PMID16390448.S2CID11872982.
- ^Tattersall, J; Rao, GV; Runac, J; Hackstadt, T; Grieshaber, SS; Grieshaber, NA (2012)."Translation inhibition of the developmental cycle protein HctA by the small RNA IhtA is conserved across Chlamydia".PLOS ONE.7(10): e47439.Bibcode:2012PLoSO...747439T.doi:10.1371/journal.pone.0047439.PMC3469542.PMID23071807.
- ^"Chlamydia protection".RetrievedAugust 1,2010.
- ^"Chlamydia".World Health Organization.17 July 2023.Retrieved2023-12-25.
- ^"Facts about chlamydia".European Centre for Disease Prevention and Control.Retrieved2023-12-25.
- ^Liechti, G. W.; Kuru, E.; Hall, E.; Kalinda, A.; Brun, Y. V.; VanNieuwenhze, M.; Maurelli, A. T. (February 2014)."A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis".Nature.506(7489): 507–510.doi:10.1038/nature12892.ISSN1476-4687.PMC3997218.PMID24336210.
- ^McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT (2006)."L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine".Proc. Natl. Acad. Sci. U.S.A.103(47): 17909–14.Bibcode:2006PNAS..10317909M.doi:10.1073/pnas.0608643103.PMC1693846.PMID17093042.
- ^"The LTP".Retrieved20 November2023.
- ^"LTP_all tree in newick format".Retrieved20 November2023.
- ^"LTP_08_2023 Release Notes"(PDF).Retrieved20 November2023.
- ^"GTDB release 08-RS214".Genome Taxonomy Database.Retrieved10 May2023.
- ^"bac120_r214.sp_label".Genome Taxonomy Database.Retrieved10 May2023.
- ^"Taxon History".Genome Taxonomy Database.Retrieved10 May2023.
Further reading
edit- Data related toChlamydiaat Wikispecies
- Chlamydiae.com