Thecytosol,also known ascytoplasmic matrixorgroundplasm,[2]is one of the liquids found inside cells (intracellular fluid(ICF)).[3]It is separated into compartments by membranes. For example, themitochondrial matrixseparates the mitochondrion into many compartments.
| Cell biology | |
|---|---|
| Animal cell diagram | |
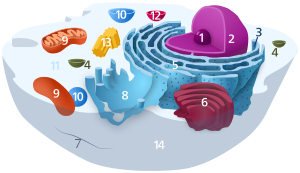 Components of a typical animal cell:
|
In theeukaryotic cell,the cytosol is surrounded by thecell membraneand is part of thecytoplasm,which also comprises the mitochondria,plastids,and otherorganelles(but not their internal fluids and structures); thecell nucleusis separate. The cytosol is thus a liquid matrix around the organelles. Inprokaryotes,most of the chemical reactions ofmetabolismtake place in the cytosol, while a few take place in membranes or in theperiplasmic space.In eukaryotes, while manymetabolic pathwaysstill occur in the cytosol, others take place within organelles.
The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and properties within cells is not well understood. The concentrations ofionssuch assodiumandpotassiumin the cytosol are different to those in theextracellular fluid;these differences in ion levels are important in processes such asosmoregulation,cell signaling,and the generation ofaction potentialsin excitable cells such as endocrine, nerve and muscle cells. The cytosol also contains large amounts ofmacromolecules,which can alter how molecules behave, throughmacromolecular crowding.
Although it was once thought to be a simple solution of molecules, the cytosol has multiple levels of organization. These includeconcentration gradientsof small molecules such ascalcium,large complexes ofenzymesthat act together and take part inmetabolic pathways,andprotein complexessuch asproteasomesandcarboxysomesthat enclose and separate parts of the cytosol.
Definition
editThe term "cytosol" was first introduced in 1965 by H. A. Lardy, and initially referred to the liquid that was produced by breaking cells apart and pelleting all the insoluble components byultracentrifugation.[4][5]Such a soluble cell extract is not identical to the soluble part of the cell cytoplasm and is usually called a cytoplasmic fraction.[6]
The termcytosolis now used to refer to the liquid phase of the cytoplasm in an intact cell.[6]This excludes any part of the cytoplasm that is contained within organelles.[7]Due to the possibility of confusion between the use of the word "cytosol" to refer to both extracts of cells and the soluble part of the cytoplasm in intact cells, the phrase "aqueous cytoplasm" has been used to describe the liquid contents of the cytoplasm of living cells.[5]
Prior to this, other terms, includinghyaloplasm,[8]were used for the cell fluid, not always synonymously, as its nature was not well understood (seeprotoplasm).[6]
Properties and composition
editThe proportion of cell volume that is cytosol varies: for example while this compartment forms the bulk of cell structure inbacteria,[9]in plant cells the main compartment is the large centralvacuole.[10]The cytosol consists mostly of water, dissolved ions, small molecules, and large water-soluble molecules (such as proteins). The majority of these non-protein molecules have amolecular massof less than 300Da.[11]This mixture of small molecules is extraordinarily complex, as the variety of molecules that are involved in metabolism (themetabolites) is immense. For example, up to 200,000 different small molecules might be made in plants, although not all these will be present in the same species, or in a single cell.[12]Estimates of the number of metabolites in single cells such asE. coliandbaker's yeastpredict that under 1,000 are made.[13][14]
Water
editMost of the cytosol iswater,which makes up about 70% of the total volume of a typical cell.[15]ThepHof the intracellular fluid is 7.4.[16]while human cytosolicpHranges between 7.0–7.4, and is usually higher if a cell is growing.[17]Theviscosityof cytoplasm is roughly the same as pure water, althoughdiffusionof small molecules through this liquid is about fourfold slower than in pure water, due mostly to collisions with the large numbers ofmacromoleculesin the cytosol.[18]Studies in thebrine shrimphave examined how water affects cell functions; these saw that a 20% reduction in the amount of water in a cell inhibits metabolism, with metabolism decreasing progressively as the cell dries out and all metabolic activity halting when the water level reaches 70% below normal.[5]
Although water is vital for life, the structure of this water in the cytosol is not well understood, mostly because methods such asnuclear magnetic resonance spectroscopyonly give information on the average structure of water, and cannot measure local variations at the microscopic scale. Even the structure of pure water is poorly understood, due to the ability of water to form structures such aswater clustersthroughhydrogen bonds.[19]
The classic view of water in cells is that about 5% of this water is strongly bound in by solutes or macromolecules as water ofsolvation,while the majority has the same structure as pure water.[5]This water of solvation is not active inosmosisand may have different solvent properties, so that some dissolved molecules are excluded, while others become concentrated.[20][21]However, others argue that the effects of the high concentrations of macromolecules in cells extend throughout the cytosol and that water in cells behaves very differently from the water in dilute solutions.[22]These ideas include the proposal that cells contain zones of low and high-density water, which could have widespread effects on the structures and functions of the other parts of the cell.[19][23]However, the use of advanced nuclear magnetic resonance methods to directly measure the mobility of water in living cells contradicts this idea, as it suggests that 85% of cell water acts like that pure water, while the remainder is less mobile and probably bound to macromolecules.[24]
Ions
editThe concentrations of the otherionsin cytosol are quite different from those inextracellular fluidand the cytosol also contains much higher amounts of charged macromolecules such as proteins and nucleic acids than the outside of the cell structure.
| Ion | Concentration(millimolar) | |
|---|---|---|
| In cytosol | In plasma | |
| Potassium | 139–150[25][26] | 4 |
| Sodium | 12 | 145 |
| Chloride | 4 | 116 |
| Bicarbonate | 12 | 29 |
| Amino acidsin proteins | 138 | 9 |
| Magnesium | 0.8 | 1.5 |
| Calcium | <0.0002 | 1.8 |
In contrast to extracellular fluid, cytosol has a high concentration ofpotassiumions and a low concentration ofsodiumions.[27]This difference in ion concentrations is critical forosmoregulation,since if the ion levels were the same inside a cell as outside, water would enter constantly byosmosis- since the levels ofmacromoleculesinside cells are higher than their levels outside. Instead, sodium ions are expelled and potassium ions taken up by theNa⁺/K⁺-ATPase,potassium ions then flow down their concentration gradient through potassium-selection ion channels, this loss of positive charge creates a negativemembrane potential.To balance thispotential difference,negative chloride ions also exit the cell, through selective chloride channels. The loss of sodium and chloride ions compensates for the osmotic effect of the higher concentration of organic molecules inside the cell.[27]
Cells can deal with even larger osmotic changes by accumulatingosmoprotectantssuch asbetainesortrehalosein their cytosol.[27]Some of these molecules can allow cells to survive being completely dried out and allow an organism to enter a state of suspended animation calledcryptobiosis.[28]In this state the cytosol and osmoprotectants become a glass-like solid that helps stabilize proteins and cell membranes from the damaging effects of desiccation.[29]
The low concentration ofcalciumin the cytosol allows calcium ions to function as asecond messengerincalcium signaling.Here, a signal such as ahormoneor anaction potentialopenscalcium channelso that calcium floods into the cytosol.[30]This sudden increase in cytosolic calcium activates other signalling molecules, such ascalmodulinandprotein kinase C.[31]Other ions such as chloride and potassium may also have signaling functions in the cytosol, but these are not well understood.[32]
Macromolecules
editProtein molecules that do not bind tocell membranesor thecytoskeletonare dissolved in the cytosol. The amount of protein in cells is extremely high, and approaches 200 mg/ml, occupying about 20–30% of the volume of the cytosol.[1]However, measuring precisely how much protein is dissolved in cytosol in intact cells is difficult, since some proteins appear to be weakly associated with membranes or organelles in whole cells and are released into solution uponcell lysis.[5]Indeed, in experiments where the plasma membrane of cells were carefully disrupted usingsaponin,without damaging the other cell membranes, only about one quarter of cell protein was released. These cells were also able to synthesize proteins if given ATP and amino acids, implying that many of the enzymes in cytosol are bound to the cytoskeleton.[33]However, the idea that the majority of the proteins in cells are tightly bound in a network called themicrotrabecular latticeis now seen as unlikely.[34]
In prokaryotes the cytosol contains the cell'sgenome,within a structure known as anucleoid.[35]This is an irregular mass ofDNAand associated proteins that control thetranscriptionandreplicationof the bacterialchromosomeandplasmids.In eukaryotes the genome is held within thecell nucleus,which is separated from the cytosol bynuclear poresthat block the free diffusion of any molecule larger than about 10nanometresin diameter.[36]
This high concentration of macromolecules in cytosol causes an effect calledmacromolecular crowding,which is when theeffective concentrationof other macromolecules is increased, since they have less volume to move in. This crowding effect can produce large changes in both theratesand the position ofchemical equilibriumof reactions in the cytosol.[1]It is particularly important in its ability to alterdissociation constantsby favoring the association of macromolecules, such as when multiple proteins come together to formprotein complexes,or whenDNA-binding proteinsbind to their targets in thegenome.[37]
Organization
editAlthough the components of the cytosol are not separated into regions by cell membranes, these components do not always mix randomly and several levels of organization can localize specific molecules to defined sites within the cytosol.[38]
Concentration gradients
editAlthough small moleculesdiffuserapidly in the cytosol, concentration gradients can still be produced within this compartment. A well-studied example of these are the "calcium sparks" that are produced for a short period in the region around an opencalcium channel.[39]These are about 2micrometresin diameter and last for only a fewmilliseconds,although several sparks can merge to form larger gradients, called "calcium waves".[40]Concentration gradients of other small molecules, such asoxygenandadenosine triphosphatemay be produced in cells around clusters ofmitochondria,although these are less well understood.[41][42]
Protein complexes
editProteins can associate to formprotein complexes,these often contain a set of proteins with similar functions, such as enzymes that carry out several steps in the same metabolic pathway.[43]This organization can allowsubstrate channeling,which is when the product of one enzyme is passed directly to the next enzyme in a pathway without being released into solution.[44]Channeling can make a pathway more rapid and efficient than it would be if the enzymes were randomly distributed in the cytosol, and can also prevent the release of unstable reaction intermediates.[45]Although a wide variety of metabolic pathways involve enzymes that are tightly bound to each other, others may involve more loosely associated complexes that are very difficult to study outside the cell.[46][47]Consequently, the importance of these complexes for metabolism in general remains unclear.
Protein compartments
editSome protein complexes contain a large central cavity that is isolated from the remainder of the cytosol. One example of such an enclosed compartment is theproteasome.[48]Here, a set of subunits form a hollow barrel containingproteasesthat degrade cytosolic proteins. Since these would be damaging if they mixed freely with the remainder of the cytosol, the barrel is capped by a set of regulatory proteins that recognize proteins with a signal directing them for degradation (aubiquitintag) and feed them into the proteolytic cavity.[49]
Another large class of protein compartments arebacterial microcompartments,which are made of a protein shell that encapsulates various enzymes.[50]These compartments are typically about 100–200nanometresacross and made of interlocking proteins.[51]A well-understood example is thecarboxysome,which contains enzymes involved incarbon fixationsuch asRuBisCO.[52]
Biomolecular condensates
editNon-membrane bound organelles can form asbiomolecular condensates,which arise by clustering,oligomerisation,orpolymerisationofmacromoleculesto drivecolloidalphase separation of the cytoplasm or nucleus.
Cytoskeletal sieving
editAlthough thecytoskeletonis not part of the cytosol, the presence of this network of filaments restricts the diffusion of large particles in the cell. For example, in several studies tracer particles larger than about 25nanometres(about the size of aribosome)[53]were excluded from parts of the cytosol around the edges of the cell and next to the nucleus.[54][55]These "excluding compartments" may contain a much denser meshwork ofactinfibres than the remainder of the cytosol. These microdomains could influence the distribution of large structures such asribosomesand organelles within the cytosol by excluding them from some areas and concentrating them in others.[56]
Function
editThe cytosol is the site of multiple cell processes. Examples of these processes includesignal transductionfrom the cell membrane to sites within the cell, such as thecell nucleus,[57]or organelles.[58]This compartment is also the site of many of the processes ofcytokinesis,after the breakdown of thenuclear membraneinmitosis.[59]Another major function of cytosol is to transport metabolites from their site of production to where they are used. This is relatively simple for water-soluble molecules, such as amino acids, which can diffuse rapidly through the cytosol.[18]However,hydrophobicmolecules, such asfatty acidsorsterols,can be transported through the cytosol by specific binding proteins, which shuttle these molecules between cell membranes.[60][61]Molecules taken into the cell byendocytosisor on their way to besecretedcan also be transported through the cytosol insidevesicles,[62]which are small spheres of lipids that are moved along the cytoskeleton bymotor proteins.[63]
The cytosol is the site of most metabolism in prokaryotes,[9]and a large proportion of the metabolism of eukaryotes. For instance, in mammals about half of the proteins in the cell are localized to the cytosol.[64]The most complete data are available in yeast, where metabolic reconstructions indicate that the majority of both metabolic processes and metabolites occur in the cytosol.[65]Major metabolic pathways that occur in the cytosol in animals areprotein biosynthesis,thepentose phosphate pathway,glycolysisandgluconeogenesis.[66]The localization of pathways can be different in other organisms, for instance fatty acid synthesis occurs inchloroplastsin plants[67][68]and inapicoplastsinapicomplexa.[69]
References
edit- ^abcEllis RJ (October 2001). "Macromolecular crowding: obvious but underappreciated".Trends Biochem. Sci.26(10): 597–604.doi:10.1016/S0968-0004(01)01938-7.PMID11590012.
- ^Cammack, Richard; Atwood, Teresa; Campbell, Peter; Parish, Howard; Smith, Anthony; Vella, Frank; Stirling, John (2006). Cammack, Richard; Atwood, Teresa; Campbell, Peter; Parish, Howard; Smith, Anthony; Vella, Frank; Stirling, John (eds.)."Cytoplasmic matrix".Oxford Dictionary of Biochemistry and Molecular Biology.Oxford University Press.doi:10.1093/acref/9780198529170.001.0001.ISBN9780198529170.
- ^Liachovitzky, Carlos (2015)."Human Anatomy and Physiology Preparatory Course"(pdf).Open Educational Resources.CUNY Academic Works: 69.Archivedfrom the original on 2017-08-23.Retrieved2021-06-22.
- ^Lardry, H. A. 1969. On the direction of pyridine nucleotide oxidation-reduction reactions in gluconeogenesis and lipogenesis. In:Control of energy metabolism,edited by B. Chance, R. Estabrook, and J. R. Williamson. New York: Academic, 1965, p. 245,[1].
- ^abcdeClegg James S.(1984). "Properties and metabolism of the aqueous cytoplasm and its boundaries".Am. J. Physiol.246(2 Pt 2): R133–51.doi:10.1152/ajpregu.1984.246.2.R133.PMID6364846.S2CID30351411.
- ^abcCammack, Richard; Teresa Atwood; Attwood, Teresa K.; Campbell, Peter Scott; Parish, Howard I.; Smith, Tony; Vella, Frank; Stirling, John (2006).Oxford dictionary of biochemistry and molecular biology.Oxford [Oxfordshire]: Oxford University Press.ISBN0-19-852917-1.OCLC225587597.
- ^abLodish, Harvey F. (1999).Molecular cell biology.New York: Scientific American Books.ISBN0-7167-3136-3.OCLC174431482.
- ^Hanstein, J. (1880).Das Protoplasma.Heidelberg. p. 24.
- ^abHoppert M, Mayer F (1999). "Principles of macromolecular organization and cell function in bacteria and archaea".Cell Biochem. Biophys.31(3): 247–84.doi:10.1007/BF02738242.PMID10736750.S2CID21004307.
- ^Bowsher CG, Tobin AK (April 2001)."Compartmentation of metabolism within mitochondria and plastids".J. Exp. Bot.52(356): 513–27.doi:10.1093/jexbot/52.356.513.PMID11373301.
- ^Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB (May 2004)."Metabolomics by numbers: acquiring and understanding global metabolite data"(PDF).Trends Biotechnol.22(5): 245–52.doi:10.1016/j.tibtech.2004.03.007.PMID15109811.Archived fromthe original(PDF)on 2008-12-17.
- ^Weckwerth W (2003). "Metabolomics in systems biology".Annu Rev Plant Biol.54:669–89.doi:10.1146/annurev.arplant.54.031902.135014.PMID14503007.S2CID1197884.
- ^Reed JL, Vo TD, Schilling CH, Palsson BO (2003)."An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR)".Genome Biol.4(9): R54.doi:10.1186/gb-2003-4-9-r54.PMC193654.PMID12952533.
- ^Förster J, Famili I, Fu P, Palsson BØ, Nielsen J (February 2003)."Genome-Scale Reconstruction of the Saccharomyces cerevisiae Metabolic Network".Genome Res.13(2): 244–53.doi:10.1101/gr.234503.PMC420374.PMID12566402.
- ^Luby-Phelps K (2000)."Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area"(PDF).Int. Rev. Cytol.International Review of Cytology.192:189–221.doi:10.1016/S0074-7696(08)60527-6.ISBN978-0-12-364596-8.PMID10553280.Archived fromthe original(PDF)on 2011-07-19.
- ^Roos A, Boron WF (April 1981). "Intracellular pH".Physiol. Rev.61(2): 296–434.doi:10.1152/physrev.1981.61.2.296.PMID7012859.
- ^Bright, G R; Fisher, GW; Rogowska, J; Taylor, DL (1987)."Fluorescence ratio imaging microscopy: temporal and spatial measurements of cytoplasmic pH".The Journal of Cell Biology.104(4): 1019–1033.doi:10.1083/jcb.104.4.1019.PMC2114443.PMID3558476.
- ^abVerkman AS (January 2002). "Solute and macromolecule diffusion in cellular aqueous compartments".Trends Biochem. Sci.27(1): 27–33.doi:10.1016/S0968-0004(01)02003-5.PMID11796221.
- ^abWiggins PM(1 December 1990)."Role of water in some biological processes".Microbiol. Rev.54(4): 432–49.doi:10.1128/MMBR.54.4.432-449.1990.PMC372788.PMID2087221.
- ^Fulton AB (September 1982). "How crowded is the cytoplasm?".Cell.30(2): 345–7.doi:10.1016/0092-8674(82)90231-8.PMID6754085.S2CID6370250.
- ^Garlid KD (2000). "The state of water in biological systems".Int. Rev. Cytol.International Review of Cytology.192:281–302.doi:10.1016/S0074-7696(08)60530-6.ISBN978-0-12-364596-8.PMID10553283.
- ^Chaplin M (November 2006). "Do we underestimate the importance of water in cell biology?".Nat. Rev. Mol. Cell Biol.7(11): 861–6.doi:10.1038/nrm2021.PMID16955076.S2CID42919563.
- ^Wiggins PM(June 1996). "High and low density water and resting, active and transformed cells".Cell Biol. Int.20(6): 429–35.doi:10.1006/cbir.1996.0054.PMID8963257.S2CID42866068.
- ^Persson E, Halle B (April 2008)."Cell water dynamics on multiple time scales".Proc. Natl. Acad. Sci. U.S.A.105(17): 6266–71.Bibcode:2008PNAS..105.6266P.doi:10.1073/pnas.0709585105.PMC2359779.PMID18436650.
- ^Thier, S. O. (April 25, 1986). "Potassium physiology".The American Journal of Medicine.80(4A): 3–7.doi:10.1016/0002-9343(86)90334-7.PMID3706350.
- ^Lote, Christopher J. (2012).Principles of Renal Physiology, 5th edition.Springer. p. 12.
- ^abcLang F (October 2007). "Mechanisms and significance of cell volume regulation".J Am Coll Nutr.26(5 Suppl): 613S–623S.doi:10.1080/07315724.2007.10719667.PMID17921474.S2CID1798009.
- ^Sussich F, Skopec C, Brady J, Cesàro A (August 2001). "Reversible dehydration of trehalose and anhydrobiosis: from solution state to an exotic crystal?".Carbohydr. Res.334(3): 165–76.doi:10.1016/S0008-6215(01)00189-6.PMID11513823.
- ^Crowe JH, Carpenter JF, Crowe LM (1998). "The role of vitrification in anhydrobiosis".Annu. Rev. Physiol.60:73–103.doi:10.1146/annurev.physiol.60.1.73.PMID9558455.
- ^Berridge MJ (1 March 1997)."Elementary and global aspects of calcium signalling".J. Physiol.499(Pt 2): 291–306.doi:10.1113/jphysiol.1997.sp021927.PMC1159305.PMID9080360.
- ^Kikkawa U, Kishimoto A, Nishizuka Y (1989). "The protein kinase C family: heterogeneity and its implications".Annu. Rev. Biochem.58:31–44.doi:10.1146/annurev.bi.58.070189.000335.PMID2549852.
- ^Orlov SN, Hamet P (April 2006). "Intracellular monovalent ions as second messengers".J. Membr. Biol.210(3): 161–72.doi:10.1007/s00232-006-0857-9.PMID16909338.S2CID26068558.
- ^Hudder A, Nathanson L, Deutscher MP (December 2003)."Organization of Mammalian Cytoplasm".Mol. Cell. Biol.23(24): 9318–26.doi:10.1128/MCB.23.24.9318-9326.2003.PMC309675.PMID14645541.
- ^Heuser J (2002). "Whatever happened to the 'microtrabecular concept'?".Biol Cell.94(9): 561–96.doi:10.1016/S0248-4900(02)00013-8.PMID12732437.S2CID45792524.
- ^Thanbichler M, Wang S, Shapiro L (2005)."The bacterial nucleoid: a highly organized and dynamic structure".J Cell Biochem.96(3): 506–21.doi:10.1002/jcb.20519.PMID15988757.S2CID25355087.
- ^Peters R (2006). "Introduction to Nucleocytoplasmic Transport".Xenopus Protocols.Methods in Molecular Biology. Vol. 322. pp. 235–58.doi:10.1007/978-1-59745-000-3_17.ISBN978-1-58829-362-6.PMID16739728.
- ^Zhou HX, Rivas G, Minton AP (2008)."Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences".Annu Rev Biophys.37:375–97.doi:10.1146/annurev.biophys.37.032807.125817.PMC2826134.PMID18573087.
- ^Norris V, den Blaauwen T, Cabin-Flaman A (March 2007)."Functional Taxonomy of Bacterial Hyperstructures".Microbiol. Mol. Biol. Rev.71(1): 230–53.doi:10.1128/MMBR.00035-06.PMC1847379.PMID17347523.
- ^Wang SQ, Wei C, Zhao G (April 2004)."Imaging microdomain Ca2+ in muscle cells".Circ. Res.94(8): 1011–22.doi:10.1161/01.RES.0000125883.68447.A1.PMID15117829.
- ^Jaffe LF (November 1993). "Classes and mechanisms of calcium waves".Cell Calcium.14(10): 736–45.doi:10.1016/0143-4160(93)90099-R.PMID8131190.
- ^Aw, T.Y. (2000). "Intracellular compartmentation of organelles and gradients of low molecular weight species".Int Rev Cytol.International Review of Cytology.192:223–53.doi:10.1016/S0074-7696(08)60528-8.ISBN978-0-12-364596-8.PMID10553281.
- ^Weiss JN, Korge P (20 July 2001)."The cytoplasm: no longer a well-mixed bag".Circ. Res.89(2): 108–10.doi:10.1161/res.89.2.108.PMID11463714.
- ^Srere PA (1987). "Complexes of sequential metabolic enzymes".Annu. Rev. Biochem.56:89–124.doi:10.1146/annurev.bi.56.070187.000513.PMID2441660.
- ^Perham RN (2000). "Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions".Annu. Rev. Biochem.69:961–1004.doi:10.1146/annurev.biochem.69.1.961.PMID10966480.
- ^Huang X, Holden HM, Raushel FM (2001). "Channeling of substrates and intermediates in enzyme-catalyzed reactions".Annu. Rev. Biochem.70:149–80.doi:10.1146/annurev.biochem.70.1.149.PMID11395405.S2CID16722363.
- ^Mowbray J, Moses V (June 1976). "The tentative identification in Escherichia coli of a multienzyme complex with glycolytic activity".Eur. J. Biochem.66(1): 25–36.doi:10.1111/j.1432-1033.1976.tb10421.x.PMID133800.
- ^Srivastava DK, Bernhard SA (November 1986). "Metabolite transfer via enzyme-enzyme complexes".Science.234(4780): 1081–6.Bibcode:1986Sci...234.1081S.doi:10.1126/science.3775377.PMID3775377.
- ^Groll M, Clausen T (December 2003). "Molecular shredders: how proteasomes fulfill their role".Curr. Opin. Struct. Biol.13(6): 665–73.doi:10.1016/j.sbi.2003.10.005.PMID14675543.
- ^Nandi D, Tahiliani P, Kumar A, Chandu D (March 2006)."The ubiquitin-proteasome system"(PDF).J. Biosci.31(1): 137–55.doi:10.1007/BF02705243.PMID16595883.S2CID21603835.Archived(PDF)from the original on 2006-07-02.
- ^Bobik, T. A. (2007)."Bacterial Microcompartments"(PDF).Microbe.2.Am Soc Microbiol: 25–31. Archived fromthe original(PDF)on 2008-08-02.
- ^Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (August 2008). "Protein-based organelles in bacteria: carboxysomes and related microcompartments".Nat. Rev. Microbiol.6(9): 681–691.doi:10.1038/nrmicro1913.PMID18679172.S2CID22666203.
- ^Badger MR, Price GD (February 2003)."CO2concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution ".J. Exp. Bot.54(383): 609–22.doi:10.1093/jxb/erg076.PMID12554704.
- ^Cate JH (November 2001)."Construction of low-resolution x-ray crystallographic electron density maps of the ribosome".Methods.25(3): 303–8.doi:10.1006/meth.2001.1242.PMID11860284.
- ^Provance DW, McDowall A, Marko M, Luby-Phelps K (1 October 1993)."Cytoarchitecture of size-excluding compartments in living cells".J. Cell Sci.106(2): 565–77.doi:10.1242/jcs.106.2.565.PMID7980739.
- ^Luby-Phelps K, Castle PE, Taylor DL, Lanni F (July 1987)."Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells".Proc. Natl. Acad. Sci. U.S.A.84(14): 4910–3.Bibcode:1987PNAS...84.4910L.doi:10.1073/pnas.84.14.4910.PMC305216.PMID3474634.
- ^Luby-Phelps K (June 1993). "Effect of cytoarchitecture on the transport and localization of protein synthetic machinery".J. Cell. Biochem.52(2): 140–7.doi:10.1002/jcb.240520205.PMID8366131.S2CID12063324.
- ^Kholodenko BN (June 2003). "Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors".J. Exp. Biol.206(Pt 12): 2073–82.doi:10.1242/jeb.00298.PMID12756289.S2CID18002214.
- ^Pesaresi P, Schneider A, Kleine T, Leister D (December 2007). "Interorganellar communication".Curr. Opin. Plant Biol.10(6): 600–6.doi:10.1016/j.pbi.2007.07.007.PMID17719262.
- ^Winey M, Mamay CL, O'Toole ET (June 1995)."Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle".J. Cell Biol.129(6): 1601–15.doi:10.1083/jcb.129.6.1601.PMC2291174.PMID7790357.
- ^Weisiger RA (October 2002). "Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands".Mol. Cell. Biochem.239(1–2): 35–43.doi:10.1023/A:1020550405578.PMID12479566.S2CID9608133.
- ^Maxfield FR, Mondal M (June 2006). "Sterol and lipid trafficking in mammalian cells".Biochem. Soc. Trans.34(Pt 3): 335–9.doi:10.1042/BST0340335.PMID16709155.
- ^Pelham HR (August 1999)."The Croonian Lecture 1999. Intracellular membrane traffic: getting proteins sorted".Philos. Trans. R. Soc. Lond. B Biol. Sci.354(1388): 1471–8.doi:10.1098/rstb.1999.0491.PMC1692657.PMID10515003.
- ^Kamal A, Goldstein LS (February 2002). "Principles of cargo attachment to cytoplasmic motor proteins".Curr. Opin. Cell Biol.14(1): 63–8.doi:10.1016/S0955-0674(01)00295-2.PMID11792546.
- ^Foster LJ, de Hoog CL, Zhang Y (April 2006)."A mammalian organelle map by protein correlation profiling".Cell.125(1): 187–99.doi:10.1016/j.cell.2006.03.022.PMID16615899.S2CID32197.
- ^Herrgård, MJ; Swainston, N; Dobson, P; Dunn, WB; Arga, KY; Arvas, M; Blüthgen, N; Borger, S; Costenoble, R; et al. (October 2008)."A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology".Nature Biotechnology.26(10): 1155–60.doi:10.1038/nbt1492.PMC4018421.PMID18846089.
- ^Stryer, Lubert; Berg, Jeremy Mark; Tymoczko, John L. (2002).Biochemistry.San Francisco: W.H. Freeman.ISBN0-7167-4684-0.OCLC179705944.
- ^Ohlrogge J, Pollard M, Bao X (December 2000). "Fatty acid synthesis: from CO2to functional genomics ".Biochem. Soc. Trans.28(6): 567–73.doi:10.1042/BST0280567.PMID11171129.
- ^Ohlrogge JB, Kuhn DN, Stumpf PK (March 1979)."Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea".Proc. Natl. Acad. Sci. U.S.A.76(3): 1194–8.Bibcode:1979PNAS...76.1194O.doi:10.1073/pnas.76.3.1194.PMC383216.PMID286305.
- ^Goodman CD, McFadden GI (January 2007). "Fatty acid biosynthesis as a drug target in apicomplexan parasites".Curr Drug Targets.8(1): 15–30.doi:10.2174/138945007779315579.PMID17266528.S2CID2565225.
Further reading
edit- Wheatley, Denys N.; Pollack, Gerald H.; Cameron, Ivan L. (2006).Water and the Cell.Berlin: Springer.ISBN1-4020-4926-9.OCLC71298997.