Ingenetics,adeletion(also calledgene deletion,deficiency,ordeletion mutation) (sign:Δ) is amutation(a genetic aberration) in which a part of achromosomeor a sequence ofDNAis left out during DNA replication. Any number ofnucleotidescan be deleted, from a singlebaseto an entire piece of chromosome.[1]Somechromosomeshave fragile spots where breaks occur, which result in thedeletionof a part of the chromosome. The breaks can be induced by heat, viruses, radiation, or chemical reactions. When achromosomebreaks, if a part of it is deleted or lost, the missing piece of chromosome is referred to as a deletion or a deficiency.[2]
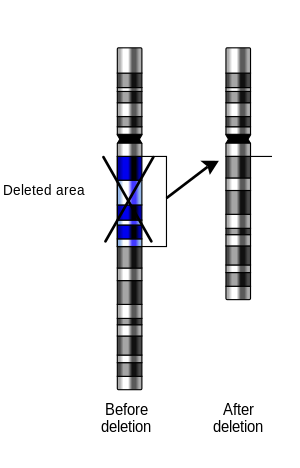
Forsynapsisto occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into adeletionorcompensation loop.
The smallest single base deletion mutations occur by a singlebase flippingin the template DNA, followed by template DNA strand slippage, within the DNA polymerase active site.[3][4][5]
Deletions can be caused by errors inchromosomal crossoverduringmeiosis,which causes several seriousgenetic diseases.Deletions that do not occur in multiples of three bases can cause aframeshiftby changing the 3-nucleotide protein reading frame of the genetic sequence. Deletions are representative ofeukaryoticorganisms, including humans and not inprokaryoticorganisms, such as bacteria.
Causes
editCauses include the following:
- Losses fromtranslocation
- Chromosomal crossoverswithin achromosomal inversion
- Unequal crossing over
- Breaking without rejoining
Types
editTypes of deletion include the following:
- Terminal deletion– a deletion that occurs towards the end of a chromosome.
- Intercalary/interstitial deletion– a deletion that occurs from the interior of a chromosome.
- Microdeletion– a relatively small amount of deletion (up to 5Mb that could include a dozen genes).
Micro-deletion is usually found in children with physical abnormalities. A large amount of deletion would result in immediate abortion (miscarriage).
Nomenclature
editTheInternational System for Human Cytogenomic Nomenclature(ISCN) is an international standard forhuman chromosomenomenclature,which includes band names, symbols and abbreviated terms used in the description of human chromosome and chromosome abnormalities. Abbreviations include a minus sign (−) for chromosome deletions, anddelfor deletions of parts of a chromosome.[7]
Effects
editSmall deletions are less likely to be fatal; large deletions are usually fatal – there are always variations based on which genes are lost. Some medium-sized deletions lead to recognizable human disorders, e.g.Williams syndrome.
Deletion of a number of pairs that is not evenly divisible by three will lead to aframeshift mutation,causing all of thecodonsoccurring after the deletion to be read incorrectly duringtranslation,producing a severely altered and potentially nonfunctionalprotein.In contrast, a deletion that is evenly divisible by three is called anin-framedeletion.[8]
Deletions are responsible for an array of genetic disorders, including some cases of maleinfertility,two thirds of cases ofDuchenne muscular dystrophy,[1]and two thirds of cases ofcystic fibrosis(those caused byΔF508).[9]Deletion of part of the short arm of chromosome 5 results inCri du chatsyndrome.[1]Deletions in theSMN-encoding gene causespinal muscular atrophy,the most common genetic cause of infant death.
Microdeletions are associated with many different conditions, including Angelman Syndrome, Prader-Willi Syndrome, and DiGeorge Syndrome.[10]Some syndromes, including Angelman syndrome and Prader-Willi syndrome, are associated with both microdeletions and genomic imprinting, meaning that same microdeletion can cause two different syndromes depending on which parent the deletion came from.[11]
Recent work suggests that some deletions of highly conserved sequences (CONDELs) may be responsible for the evolutionary differences present among closely related species. Such deletions in humans, referred to ashCONDELs,may be responsible for the anatomical and behavioral differences between humans,chimpanzeesand other varieties of mammals likeapeormonkeys.[12]
Recent comprehensive patient-level classification and quantification of driver events inTCGAcohorts revealed that there are on average 12 driver events per tumor, of which 2.1 are deletions oftumor suppressors.[13]
Detection
editThe introduction of molecular techniques in conjunction with classical cytogenetic methods has in recent years greatly improved the diagnostic potential for chromosomal abnormalities. In particular, microarray-comparative genomic hybridization (CGH) based on the use of BAC clones promises a sensitive strategy for the detection of DNA copy-number changes on a genome-wide scale. The resolution of detection could be as high as >30,000 "bands" and the size of chromosomal deletion detected could as small as 5–20 kb in length.[14]Other computation methods were selected to discover DNA sequencing deletion errors such asend-sequence profiling.[15][16]
Mitochondrial DNA deletions
editIn the yeastSaccharomyces cerevisiae,the nuclear genesRad51p,Rad52p andRad59pencode proteins that are necessary for recombinational repair and are employed in the repair ofdouble strand breaksinmitochondrial DNA.[17]Loss of these proteins decreases the rate of spontaneous DNA deletion events in mitochondria.[17]This finding implies that the repair of DNA double-strand breaks by homologous recombination is a step in the formation of mitochondrial DNA deletions.
See also
editReferences
edit- ^abcLewis, R. (2004).Human Genetics: Concepts and Applications(6th ed.). McGraw Hill.ISBN978-0072951745.
- ^Klug, William S. (2015).Concepts of genetics.Michael R. Cummings, Charlotte A. Spencer, Michael Angelo Palladino (Eleventh ed.). Boston.ISBN978-0-321-94891-5.OCLC880404074.
{{cite book}}:CS1 maint: location missing publisher (link) - ^Banavali, Nilesh K. (2013). "Partial Base Flipping is Sufficient for Strand Slippage near DNA Duplex Termini".Journal of the American Chemical Society.135(22): 8274–8282.doi:10.1021/ja401573j.PMID23692220.
- ^Banavali, Nilesh K. (2013). "Analyzing the Relationship between Single Base Flipping and Strand Slippage near DNA Duplex Termini".The Journal of Physical Chemistry B.117(46): 14320–14328.doi:10.1021/jp408957c.PMID24206351.
- ^Manjari, Swati R.; Pata, Janice D.; Banavali, Nilesh K. (2014)."Cytosine Unstacking and Strand Slippage at an Insertion–Deletion Mutation Sequence in an Overhang-Containing DNA Duplex".Biochemistry.53(23): 3807–3816.doi:10.1021/bi500189g.PMC4063443.PMID24854722.
- ^Warrender JD, Moorman AV, Lord P (2019)."A fully computational and reasonable representation for karyotypes".Bioinformatics.35(24): 5264–5270.doi:10.1093/bioinformatics/btz440.PMC6954653.PMID31228194.
{{cite journal}}:CS1 maint: multiple names: authors list (link)
- "This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/) " - ^"ISCN Symbols and Abbreviated Terms".Coriell Institute for Medical Research.Retrieved2022-10-27.
- ^LSDB — Controlled vocabulary termsArchived2011-10-06 at theWayback Machineat The GEN2PHEN Knowledge Centre. Posted Fri, 08/01/2010.
- ^Mitchell, Richard Sheppard; Kumar, Vinay; Robbins, Stanley L.; Abbas, Abul K.; Fausto, Nelson (2007).Robbins basic pathology.Saunders/Elsevier.ISBN978-1-4160-2973-1.
- ^Srour, Myriam; Shevell, Michael (2015-01-01), Rosenberg, Roger N.; Pascual, Juan M. (eds.),"Chapter 14 - Global Developmental Delay and Intellectual Disability",Rosenberg's Molecular and Genetic Basis of Neurological and Psychiatric Disease (Fifth Edition),Boston: Academic Press, pp. 151–161,ISBN978-0-12-410529-4,retrieved2022-01-07
- ^Kalsner, Louisa; Chamberlain, Stormy J. (April 22, 2015)."Prader-Willi, Angelman, and 15q11-q13 duplication syndromes".Pediatric Clinics of North America.62(3): 587–606.doi:10.1016/j.pcl.2015.03.004.ISSN0031-3955.PMC4449422.PMID26022164.
- ^McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, Wenger AM, Bejerano G, Kingsley DM (March 2011)."Human-specific loss of regulatory DNA and the evolution of human-specific traits".Nature.471(7337): 216–9.Bibcode:2011Natur.471..216M.doi:10.1038/nature09774.PMC3071156.PMID21390129.
- ^Vyatkin, Alexey D.; Otnyukov, Danila V.; Leonov, Sergey V.; Belikov, Aleksey V. (14 January 2022)."Comprehensive patient-level classification and quantification of driver events in TCGA PanCanAtlas cohorts".PLOS Genetics.18(1): e1009996.doi:10.1371/journal.pgen.1009996.PMC8759692.PMID35030162.
- ^Ren, H (May 2005)."BAC-based PCR fragment microarray: high-resolution detection of chromosomal deletion and duplication breakpoints".Human Mutation.25(5): 476–482.doi:10.1002/humu.20164.PMID15832308.S2CID28030180.
- ^Shmilovici, A.; Ben-Gal, I. (2007)."Using a VOM Model for Reconstructing Potential Coding Regions in EST Sequences"(PDF).Journal of Computational Statistics.22(1): 49–69.doi:10.1007/s00180-007-0021-8.S2CID2737235.Archived fromthe original(PDF)on 2020-05-31.Retrieved2014-01-10.
- ^Volik, S.; Zhao, S.; Chin, K.; Brebner, J. H.; Herndon, D. R.; Tao, Q.; Kowbel, D.; Huang, G.; Lapuk, A.; Kuo, W.-L.; Magrane, G.; de Jong, P.; Gray, J. W.; Collins, C. (4 June 2003)."End-sequence profiling: Sequence-based analysis of aberrant genomes".Proceedings of the National Academy of Sciences.100(13): 7696–7701.Bibcode:2003PNAS..100.7696V.doi:10.1073/pnas.1232418100.PMC164650.PMID12788976.
- ^abIvanetich, K. M.; Lucas, S.; Marsh, J. A.; Ziman, M. R.; Katz, I. D.; Bradshaw, J. J. (1978). "Organic compounds. Their interaction with and degradation of hepatic microsomal drug-metabolizing enzymes in vitro".Drug Metabolism and Disposition: The Biological Fate of Chemicals.6(3): 218–225.PMID26540.