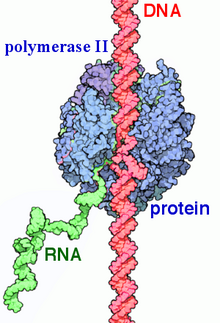
RNA polymerase II(RNAP IIandPol II) is amultiprotein complexthattranscribesDNAinto precursors ofmessenger RNA(mRNA) and mostsmall nuclear RNA(snRNA) andmicroRNA.[1][2]It is one of the threeRNAPenzymesfound in the nucleus ofeukaryoticcells.[3]A 550kDacomplex of 12 subunits, RNAP II is the most studied type ofRNA polymerase.A wide range oftranscription factorsare required for it to bind to upstream genepromotersand begin transcription.
Discovery
editEarly studies suggested a minimum of two RNAPs: one which synthesized rRNA in thenucleolus,and one which synthesized other RNA in thenucleoplasm,part of the nucleus but outside the nucleolus.[5]In 1969, biochemistsRobert G. RoederandWilliam Rutterdiscovered there are total three distinct nuclearRNA polymerases,an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm.[6]The finding was obtained by the use ofion-exchange chromatographyviaDEAEcoatedSephadexbeads. The technique separated the enzymes by the order of the corresponding elutions, Ι,ΙΙ,ΙΙΙ, by increasing the concentration of ammonium sulfate. The enzymes were named according to the order of the elutions,RNAP I,RNAP II,RNAP IΙI.[3]This discovery demonstrated that there was an additional enzyme present in the nucleoplasm, which allowed for the differentiation between RNAP II and RNAP III.[7]
RNA polymerase II (RNAP2) undergoes regulated transcriptional pausing during early elongation. Various studies has shown that disruption of transcription elongation is implicated incancer,neurodegeneration,HIV latency etc.[8]
Subunits
editTheeukaryoticcore RNA polymerase II was first purified using transcription assays.[10]The purified enzyme has typically 10–12 subunits (12 in humans and yeast) and is incapable of specific promoter recognition.[11]Many subunit-subunit interactions are known.[12]
- DNA-directed RNA polymerase II subunit RPB1– anenzymethat in humans is encoded by thePOLR2Ageneand in yeast is encoded by RPO21. RPB1 is the largest subunit of RNA polymerase II. It contains acarboxy terminal domain(CTD) composed of up to 52 heptapeptide repeats (YSPTSPS) that are essential for polymerase activity.[13]The CTD was first discovered in the laboratory of C.J. Ingles at the University of Toronto and by JL Corden atJohns Hopkins University.In combination with several other polymerase subunits, the RPB1 subunit forms the DNA binding domain of the polymerase, a groove in which the DNA template istranscribedinto RNA.[14]It strongly interacts with RPB8.[12]
- RPB2(POLR2B) – the second-largest subunit that in combination with at least two other polymerase subunits forms a structure within the polymerase that maintains contact in the active site of the enzyme between the DNA template and the newly synthesized RNA.[15]
- RPB3(POLR2C) – the third-largest subunit. Exists as a heterodimer with another polymerase subunit,POLR2Jforming a core subassembly. RPB3 strongly interacts with RPB1-5, 7, 10–12.[12]
- RNA polymerase II subunit B4(RPB4)– encoded by the POLR2Dgene[16]is the fourth-largest subunit and may have a stress protective role.
- RPB5– In humans is encoded by thePOLR2Egene. Two molecules of this subunit are present in each RNA polymerase II.[17]RPB5 strongly interacts with RPB1, RPB3, and RPB6.[12]
- RPB6 (POLR2F)– forms a structure with at least two other subunits that stabilizes the transcribing polymerase on the DNA template.[18]
- RPB7– encoded byPOLR2Gand may play a role in regulating polymerase function.[19]RPB7 interacts strongly with RPB1 and RPB5.[12]
- RPB8 (POLR2H)– interacts with subunits RPB1-3, 5, and 7.[12]
- RPB9– The groove in which the DNA template is transcribed into RNA is composed of RPB9 (POLR2I) and RPB1.
- RPB10– the product of genePOLR2L.It interacts with RPB1-3 and 5, and strongly with RPB3.[12]
- RPB11– the RPB11 subunit is itself composed of three subunits in humans:POLR2J(RPB11-a),POLR2J2(RPB11-b), and POLR2J3[20](RPB11-c).
- RPB12– Also interacts with RPB3 is RPB12 (POLR2K).[12]
Assembly
editRPB3 is involved in RNA polymerase II assembly.[21]A subcomplex of RPB2 and RPB3 appears soon after subunit synthesis.[21]This complex subsequently interacts with RPB1.[21]RPB3, RPB5, and RPB7 interact with themselves to form homodimers, and RPB3 and RPB5 together are able to contact all of the other RPB subunits, except RPB9.[12]Only RPB1 strongly binds to RPB5.[12]The RPB1 subunit also contacts RPB7, RPB10, and more weakly but most efficiently with RPB8.[12]Once RPB1 enters the complex, other subunits such as RPB5 and RPB7 can enter, where RPB5 binds to RPB6 and RPB8 and RPB3 brings in RPB10, RPB 11, and RPB12.[12]RPB4 and RPB9 may enter once most of the complex is assembled. RPB4 forms a complex with RPB7.[12]
Kinetics
editEnzymescan catalyze up to several million reactions per second. Enzyme rates depend on solution conditions and substrate concentration. Like other enzymes POLR2 has a saturation curve and a maximum velocity (Vmax). It has aKm(substrate concentration required for one-halfVmax) and akcat(the number of substrate molecules handled by one active site per second). The specificity constant is given bykcat/Km.The theoretical maximum for the specificity constant is the diffusion limit of about 108to 109(M−1s−1), where every collision of the enzyme with its substrate results in catalysis. In yeast, mutation in the Trigger-Loop domain of the largest subunit can change the kinetics of the enzyme.[22]
Bacterial RNA polymerase, a relative of RNA Polymerase II, switches between inactivated and activated states by translocating back and forth along the DNA.[23]Concentrations of [NTP]eq= 10 μM GTP, 10 μM UTP, 5 μM ATP and 2.5 μM CTP, produce a mean elongation rate, turnover number, of ~1 bp (NTP)−1for bacterial RNAP, a relative of RNA polymerase II.[23]
RNA polymerase II undergoes extensive co-transcriptional pausing during transcription elongation.[24][25]This pausing is especially pronounced at nucleosomes, and arises in part through the polymerase entering a transcriptionally incompetent backtracked state.[24]The duration of these pauses ranges from seconds to minutes or longer, and exit from long-lived pauses can be promoted by elongation factors such as TFIIS.[26]In turn, the transcription rate influences whether the histones of transcribed nucleosomes are evicted from chromatin, or reinserted behind the transcribing polymerase.[27]
Alpha-Amanitin
editRNA polymerase II is inhibited byα-Amanitin[28]and otheramatoxins.α-Amanitin is a highly poisonous substance found in many mushrooms.[5]The mushroom poison has different effects on each of the RNA Polymerases: I, II, III. RNAP I is completely unresponsive to the substance and will function normally while RNAP III has a moderate sensitivity. RNAP II, however, is completely inhibited by the toxin. Alpha-Amanitin inhibits RNAP II by strong interactions in the enzyme's "funnel", "cleft", and the key "bridgeα-helix"regions of the RPB-1 subunit.[29]
Holoenzyme
editRNA polymerase II holoenzymeis a form ofeukaryoticRNA polymerase II that is recruited to the promoters ofprotein-coding genes in living cells.[11]It consists of RNA polymerase II, a subset of generaltranscription factors,andregulatory proteinsknown as SRB proteins.
Part of the assembly of the holoenzyme is referred to as thepreinitiation complex,because its assembly takes place on thegenepromoterbefore the initiation oftranscription.Themediator complexacts as a bridge between RNA polymerase II and the transcription factors.
Control by chromatin structure
editThis is an outline of an example mechanism of yeast cells by whichchromatinstructure andhistonepost-translational modificationhelp regulate and record thetranscriptionofgenesby RNA polymerase II.
This pathway gives examples of regulation at these points of transcription:
- Pre-initiation (promotion by Bre1, histone modification)
- Initiation (promotion by TFIIH, Pol II modification and promotion by COMPASS, histone modification)
- Elongation (promotion by Set2, Histone Modification)
This refers to various stages of the process as regulatory steps. It has not been proven that they are used for regulation, but is very likely they are.
RNA Pol II elongation promoters can be summarised in 3 classes.
- Drug/sequence-dependent arrest-affected factors (Various interfering proteins)
- Chromatin structure-oriented factors (Histone posttranscriptional modifiers, e.g., Histone Methyltransferases)
- RNA Pol II catalysis-improving factors (Various interfering proteins and Pol II cofactors; see RNA polymerase II).
Transcription mechanisms
edit- Chromatin structure oriented factors:
(HMTs (HistoneMethylTransferases)):
COMPASS§† – (COMplex ofProteinsASsociated withSet1) – Methylates lysine 4 of histone H3: Is responsible of repression/silencing of transcription. A normal part of cell growth and transcription regulation within RNAP II.[30] - Set2 – Methylates lysine 36 of histone H3: Set2 is involved in regulation transcription elongation through its direct contact with the CTD.[31]
(interesting irrelevant example: Dot1*‡ – Methylates lysine 79 of histone H3.) - Bre1 – Ubiquinates (addsubiquitinto) lysine 123 of histone H2B. Associated with pre-initiation and allowing RNA Pol II binding.
C-terminal Domain
editTheC-terminusof RPB1 is appended to form the C-terminal domain (CTD). The carboxy-terminal domain of RNA polymerase II typically consists of up to 52 repeats of the sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser.[32]The domain stretches from the core of the RNAPII enzyme to the exit channel, this placement is effective due to its inductions of "RNA processing reactions, through direct or indirect interactions with components of the RNA processing machinery".[33]The CTD domain does not exist in RNA Polymerase I or RNA Polymerase III.[3]The RNA Polymerase CTD was discovered first in the laboratory of C. J. Ingles at the University of Toronto and also in the laboratory of J Corden at Johns Hopkins University during the processes of sequencing the DNA encoding the RPB1 subunit of RNA polymerase from yeast and mice respectively. Other proteins often bind the C-terminal domain of RNA polymerase in order to activate polymerase activity. It is the protein domain that is involved in theinitiationof transcription, thecappingof theRNA transcript,and attachment to thespliceosomeforRNA splicing.[13]
Phosphorylation of the CTD
editRNA Polymerase II exists in two forms unphosphorylated and phosphorylated, IIA and IIO respectively.[5][3]The transition between the two forms facilitates different functions for transcription. Thephosphorylationof CTD is catalyzed by one of the sixgeneral transcription factors,TFIIH.TFIIH serves two purposes: one is to unwind the DNA at the transcription start site and the other is to phosphorylate. The form polymerase IIA joins the preinitiation complex, this is suggested because IIA binds with higher affinity to the TBP (TATA-box binding protein), the subunit of the general transcription factorTFIID,than polymerase IIO form. The form polymerase IIO facilitates the elongation of the RNA chain.[5]The method for the elongation initiation is done by the phosphorylation ofserineat position 5 (Ser5), via TFIIH. The newly phosphorylated Ser5 recruits enzymes to cap the 5' end of the newly synthesized RNA and the "3' processing factors topoly(A)sites ".[33]Once the second serine is phosphorylated, Ser2, elongation is activated. In order to terminate elongation dephosphorylation must occur. Once the domain is completely dephosphorylated the RNAP II enzyme is "recycled" and catalyzes the same process with another initiation site.[33]
Transcription coupled recombinational repair
editOxidative DNA damagemay block RNA polymerase IItranscriptionand cause strand breaks. An RNA templated transcription-associated recombination process has been described that can protect against DNA damage.[34]During the G1/G0 stages of thecell cycle,cells exhibit assembly of homologous recombination factors at double-strand breaks within actively transcribed regions. It appears that transcription is coupled to repair of DNA double-strand breaks by RNA templated homologous recombination. This repair process efficiently and accurately rejoins double-strand breaks ingenesbeing actively transcribed by RNA polymerase II.
See also
editReferences
edit- ^Kornberg RD (December 1999)."Eukaryotic transcriptional control".Trends in Cell Biology.9(12): M46–9.doi:10.1016/S0962-8924(99)01679-7.PMID10611681.
- ^Sims RJ, Mandal SS, Reinberg D (June 2004)."Recent highlights of RNA-polymerase-II-mediated transcription".Current Opinion in Cell Biology.16(3): 263–71.doi:10.1016/j.ceb.2004.04.004.PMID15145350.
- ^abcdYoung, Richard A. (2003-11-28). "RNA Polymerase II".Annual Review of Biochemistry.60(1): 689–715.doi:10.1146/annurev.bi.60.070191.003353.PMID1883205.
- ^Meyer PA, Ye P, Zhang M, Suh MH, Fu J (June 2006)."Phasing RNA polymerase II using intrinsically bound Zn atoms: an updated structural model".Structure.14(6): 973–82.doi:10.1016/j.str.2006.04.003.PMID16765890.
- ^abcdWeaver, Robert Franklin (2012-01-01).Molecular biology.McGraw-Hill.ISBN9780073525327.OCLC789601172.
- ^Roeder RG, Rutter WJ (Oct 1969). "Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms".Nature.224(5216): 234–7.Bibcode:1969Natur.224..234R.doi:10.1038/224234a0.PMID5344598.S2CID4283528.
- ^Roeder RG, Rutter WJ (Oct 1969). "Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms".Nature.224(5216): 234–7.Bibcode:1969Natur.224..234R.doi:10.1038/224234a0.PMID5344598.S2CID4283528.
- ^Cermakova, Katerina; Demeulemeester, Jonas; Lux, Vanda; Nedomova, Monika; Goldman, Seth R.; Smith, Eric A.; Srb, Pavel; Hexnerova, Rozalie; Fabry, Milan; Madlikova, Marcela; Horejsi, Magdalena (2021-11-26)."A ubiquitous disordered protein interaction module orchestrates transcription elongation".Science.374(6571): 1113–1121.Bibcode:2021Sci...374.1113C.doi:10.1126/science.abe2913.PMC8943916.PMID34822292.S2CID244660781.
- ^Armache, Karim-Jean; Mitterweger, Simone; Meinhart, Anton; Cramer, Patrick (2019)."Structures of complete RNA polymerase II and its subcomplex, Rpb4/7"(PDF).Journal of Biological Chemistry.280(8): 7131–1734.doi:10.2210/pdb1wcm/pdb.PMID15591044.
- ^Sawadogo M, Sentenac A (1990). "RNA polymerase B (II) and general transcription factors".Annual Review of Biochemistry.59:711–54.doi:10.1146/annurev.bi.59.070190.003431.PMID2197989.
- ^abMyer VE, Young RA (October 1998)."RNA polymerase II holoenzymes and subcomplexes".The Journal of Biological Chemistry.273(43): 27757–60.doi:10.1074/jbc.273.43.27757.PMID9774381.
- ^abcdefghijklmAcker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M (July 1997)."Interactions between the human RNA polymerase II subunits".The Journal of Biological Chemistry.272(27): 16815–21.doi:10.1074/jbc.272.27.16815.PMID9201987.
- ^abBrickey WJ, Greenleaf AL (June 1995)."Functional studies of the carboxy-terminal repeat domain of Drosophila RNA polymerase II in vivo".Genetics.140(2): 599–613.doi:10.1093/genetics/140.2.599.PMC1206638.PMID7498740.
- ^"Entrez Gene: POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa".
- ^"Entrez Gene: POLR2B polymerase (RNA) II (DNA directed) polypeptide B, 140kDa".
- ^Khazak V, Estojak J, Cho H, Majors J, Sonoda G, Testa JR, Golemis EA (April 1998)."Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II".Molecular and Cellular Biology.18(4): 1935–45.doi:10.1128/mcb.18.4.1935.PMC121423.PMID9528765.
- ^"Entrez Gene: POLR2E polymerase (RNA) II (DNA directed) polypeptide E, 25kDa".
- ^"Entrez Gene: POLR2F polymerase (RNA) II (DNA directed) polypeptide F".
- ^"Entrez Gene: POLR2G polymerase (RNA) II (DNA directed) polypeptide G".
- ^"POLR2J3 polymerase (RNA) II (DNA directed) polypeptide J3".
- ^abcKolodziej PA, Young RA (September 1991)."Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly".Molecular and Cellular Biology.11(9): 4669–78.doi:10.1128/mcb.11.9.4669.PMC361357.PMID1715023.
- ^Kaplan CD, Jin H, Zhang IL, Belyanin A (April 12, 2012)."Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo".PLOS Genetics.8(4): e1002627.doi:10.1371/journal.pgen.1002627.PMC3325174.PMID22511879.
- ^abAbbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM (November 2005)."Direct observation of base-pair stepping by RNA polymerase".Nature.438(7067): 460–5.Bibcode:2005Natur.438..460A.doi:10.1038/nature04268.PMC1356566.PMID16284617.
- ^abHodges, Courtney; Bintu, Lacramioara; Lubkowska, Lucyna; Kashlev, Mikhail; Bustamante, Carlos (2009-07-31)."Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II".Science.325(5940): 626–628.Bibcode:2009Sci...325..626H.doi:10.1126/science.1172926.ISSN1095-9203.PMC2775800.PMID19644123.
- ^Churchman, L. Stirling; Weissman, Jonathan S. (2011-01-20)."Nascent transcript sequencing visualizes transcription at nucleotide resolution".Nature.469(7330): 368–373.Bibcode:2011Natur.469..368C.doi:10.1038/nature09652.ISSN1476-4687.PMC3880149.PMID21248844.
- ^Galburt, Eric A.; Grill, Stephan W.; Wiedmann, Anna; Lubkowska, Lucyna; Choy, Jason; Nogales, Eva; Kashlev, Mikhail; Bustamante, Carlos (2007-04-12). "Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner".Nature.446(7137): 820–823.Bibcode:2007Natur.446..820G.doi:10.1038/nature05701.ISSN1476-4687.PMID17361130.S2CID4310108.
- ^Bintu, Lacramioara; Kopaczynska, Marta; Hodges, Courtney; Lubkowska, Lucyna; Kashlev, Mikhail; Bustamante, Carlos (2011-11-13)."The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes".Nature Structural & Molecular Biology.18(12): 1394–1399.doi:10.1038/nsmb.2164.ISSN1545-9985.PMC3279329.PMID22081017.
- ^Kaplan CD, Larsson KM, Kornberg RD (June 2008)."The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin".Molecular Cell.30(5): 547–56.doi:10.1016/j.molcel.2008.04.023.PMC2475549.PMID18538653.
- ^Gong, Xue Q.; Nedialkov, Yuri A.; Burton, Zachary F. (2004-06-25)."α-Amanitin Blocks Translocation by Human RNA Polymerase II".Journal of Biological Chemistry.279(26): 27422–27427.doi:10.1074/jbc.M402163200.ISSN0021-9258.PMID15096519.
- ^Briggs, Scott D.; Bryk, Mary; Strahl, Brian D.; Cheung, Wang L.; Davie, Judith K.; Dent, Sharon Y. R.; Winston, Fred; Allis, C. David (2001-12-15)."Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae".Genes & Development.15(24): 3286–3295.doi:10.1101/gad.940201.ISSN0890-9369.PMC312847.PMID11751634.
- ^Li, Bing; Howe, LeAnn; Anderson, Scott; Yates, John R.; Workman, Jerry L. (2003-03-14)."The Set2 Histone Methyltransferase Functions through the Phosphorylated Carboxyl-terminal Domain of RNA Polymerase II".Journal of Biological Chemistry.278(11): 8897–8903.doi:10.1074/jbc.M212134200.ISSN0021-9258.PMID12511561.
- ^Meinhart A, Cramer P (July 2004)."Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors".Nature.430(6996): 223–6.Bibcode:2004Natur.430..223M.doi:10.1038/nature02679.hdl:11858/00-001M-0000-0015-8512-8.PMID15241417.S2CID4418258.
- ^abcEgloff, Sylvain; Murphy, Shona (2008). "Cracking the RNA polymerase II CTD code".Trends in Genetics.24(6): 280–288.doi:10.1016/j.tig.2008.03.008.PMID18457900.
- ^Wei L, Levine AS, Lan L (2016). "Transcription-coupled homologous recombination after oxidative damage".DNA Repair (Amst.).44:76–80.doi:10.1016/j.dnarep.2016.05.009.PMID27233112.
External links
edit- More information at Berkeley National Lab(Wayback Machinecopy)
- RNA+Polymerase+IIat the U.S. National Library of MedicineMedical Subject Headings(MeSH)