Thesodium–potassium pump(sodium–potassiumadenosinetriphosphatase,also known asNa+/K+-ATPase,Na+/K+pump,orsodium–potassium ATPase) is anenzyme(anelectrogenictransmembraneATPase) found in themembraneof allanimalcells. It performs several functions incell physiology.
| Na+/K+-ATPase pump | |||||||||
|---|---|---|---|---|---|---|---|---|---|
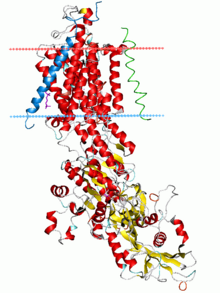 Sodium–potassium pump, E2-Pi state. Calculated hydrocarbon boundaries of thelipid bilayerare shown as blue (intracellular) and red (extracellular) planes | |||||||||
| Identifiers | |||||||||
| EC no. | 7.2.2.13 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDBstructures | RCSB PDBPDBePDBsum | ||||||||
| |||||||||
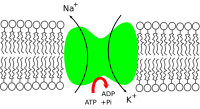

TheNa+/K+-ATPase enzyme isactive(i.e. it uses energy fromATP). For every ATP molecule that the pump uses, three sodium ions are exported and two potassium ions are imported.[1]Thus, there is a net export of a single positive charge per pump cycle. The net effect is an extracellular concentration of sodium ions which is 5 times the intracellular concentration, and an intracellular concentration of potassium ions which is 30 times the extracellular concentration.[1]
The sodium–potassium pump was discovered in 1957 by the Danish scientistJens Christian Skou,who was awarded a Nobel Prize for his work in 1997. Its discovery marked an important step forward in the understanding of how ions get into and out of cells, and it has particular significance for excitable cells such asnerve cells,which depend on this pump to respond to stimuli and transmit impulses.
All mammals have four different sodium pump sub-types, or isoforms. Each has unique properties and tissue expression patterns.[2]This enzyme belongs to the family ofP-type ATPases.
Function
editTheNa+/K+-ATPase helps maintainresting potential,affects transport, and regulates cellularvolume.[3]It also functions as a signal transducer/integrator to regulate theMAPK pathway,reactive oxygen species(ROS), as well as intracellular calcium. In fact, all cells expend a large fraction of the ATP they produce (typically 30% and up to 70% in nerve cells) to maintain their required cytosolic Na and K concentrations.[4] For neurons, theNa+/K+-ATPase can be responsible for up to 3/4 of the cell's energy expenditure.[5]In many types of tissue, ATP consumption by theNa+/K+-ATPases have been related toglycolysis.This was first discovered in red blood cells (Schrier, 1966), but has later been evidenced in renal cells,[6]smooth muscles surrounding the blood vessels,[7]andcardiac Purkinje cells.[8]Recently, glycolysis has also been shown to be of particular importance forNa+/K+-ATPase in skeletal muscles, where inhibition ofglycogenbreakdown (a substrate forglycolysis) leads to reducedNa+/K+-ATPase activity and lower force production.[9][10][11]
Resting potential
editIn order to maintain the cell membrane potential, cells keep a low concentration of sodium ions and high levels of potassium ions within the cell (intracellular). The sodium–potassium pump mechanism moves 3 sodium ions out and moves 2 potassium ions in, thus, in total, removing one positive charge carrier from theintracellular space(see§ Mechanismfor details). In addition, there is a short-circuit channel (i.e. a highly K-permeable ion channel) for potassium in the membrane, thus the voltage across the plasma membrane is close to theNernst potentialof potassium.
Reversal potential
editEven if bothK+andNa+ions have the same charge, they can still have very different equilibrium potentials for both outside and/or inside concentrations. The sodium-potassium pump moves toward a nonequilibrium state with the relative concentrations ofNa+andK+for both inside and outside of cell. For instance, the concentration ofK+in cytosol is 100mM,whereas the concentration ofNa+is 10 mM. On the other hand, in extracellular space, the usual concentration range ofK+is about 3.5-5 mM, whereas the concentration ofNa+is about 135-145 mM.[citation needed]
Transport
editExport of sodium ions from the cell provides the driving force for several secondary active transporters such asmembrane transport proteins,which importglucose,amino acidsand other nutrients into the cell by use of the sodium ion gradient.
Another important task of theNa+-K+pump is to provide aNa+gradient that is used by certain carrier processes. In thegut,for example, sodium is transported out of the reabsorbing cell on the blood (interstitial fluid) side via theNa+-K+pump, whereas, on the reabsorbing (lumenal) side, theNa+-glucosesymporteruses the createdNa+gradient as a source of energy to import bothNa+and glucose, which is far more efficient than simple diffusion. Similar processes are located in therenal tubular system.
Controlling cell volume
editFailure of theNa+-K+pumps can result in swelling of the cell. A cell'sosmolarityis the sum of the concentrations of the variousionspecies and manyproteinsand other organic compounds inside the cell. When this is higher than theosmolarityoutside of the cell, water flows into the cell throughosmosis.This can cause the cell to swell up andlyse.TheNa+-K+pump helps to maintain the right concentrations of ions. Furthermore, when the cell begins to swell, this automatically activates theNa+-K+pump because it changes the internal concentrations ofNa+-K+to which the pump is sensitive.[12]
Functioning as signal transducer
editWithin the last decade[when?],many independent labs have demonstrated that, in addition to the classical ion transporting, this membrane protein can also relay extracellularouabain-binding signalling into the cell through regulation ofprotein tyrosine phosphorylation.For instance, a study investigated the function ofNa+/K+-ATPase in foot muscle and hepatopancreas in land snailOtala lacteaby comparing the active and estivating states.[13]They concluded that reversible phosphorylation can control the same means of coordinating ATP use by this ion pump with the rates of the ATP generation by catabolic pathways in estivatingO. lactea.The downstream signals through ouabain-triggered protein phosphorylation events include activation of themitogen-activated protein kinase(MAPK) signal cascades, mitochondrialreactive oxygen species(ROS) production, as well as activation ofphospholipase C(PLC) andinositol triphosphate(IP3) receptor (IP3R) in different intracellular compartments.[14]
Protein-protein interactions play a very important role inNa+-K+pump-mediated signal transduction. For example, theNa+-K+pump interacts directly withSrc,anon-receptor tyrosine kinase,to form a signaling receptor complex.[15]Src is initially inhibited by theNa+-K+pump. However, upon subsequent ouabain binding, the Src kinase domain is released and then activated. Based on this scenario, NaKtide, a peptide Src inhibitor derived from theNa+-K+pump, was developed as a functional ouabain–Na+-K+pump-mediated signal transduction.[16]Na+-K+pump also interacts withankyrin,IP3R,PI3K,PLCgamma1andcofilin.[17]
Controlling neuron activity states
editTheNa+-K+pump has been shown to control and set the intrinsic activity mode ofcerebellarPurkinje neurons,[18]accessory olfactory bulbmitral cells[19]and probably other neuron types.[20]This suggests that the pump might not simply be ahomeostatic,"housekeeping" molecule for ionic gradients, but could be acomputationelement in thecerebellumand thebrain.[21]Indeed, amutationin theNa+-K+pump causes rapid onsetdystonia-parkinsonism,which has symptoms to indicate that it is a pathology of cerebellar computation.[22]Furthermore, anouabainblock ofNa+-K+pumps in the cerebellum of a live mouse results in it displayingataxiaanddystonia.[23]Alcoholinhibits sodium–potassium pumps in the cerebellum and this is likely how it corrupts cerebellar computation and body coordination.[24][25]The distribution of theNa+-K+pump on myelinated axons in the human brain has been demonstrated to be along the internodalaxolemma,and not within the nodal axolemma as previously thought.[26]TheNa+-K+pump disfunction has been tied to various diseases, including epilepsy and brain malformations.[27]
Mechanism
editLooking at the process starting from the interior of the cell:
- The pump has a higher affinity forNa+ions thanK+ions, thus after bindingATP,binds 3 intracellularNa+ions.[3]
- ATP ishydrolyzed,leading tophosphorylationof the pump at a highly conservedaspartateresidue and subsequent release ofADP.This process leads to a conformational change in the pump.
- The conformational change exposes theNa+ions to the extracellular region. The phosphorylated form of the pump has a low affinity forNa+ions, so they are released; by contrast it has high affinity for theK+ions.
- The pump binds 2 extracellularK+ions, which inducesdephosphorylationof the pump, reverting it to its previous conformational state, thus releasing theK+ions into the cell.
- The unphosphorylated form of the pump has a higher affinity forNa+ions. ATP binds, and the process starts again.
Regulation
editEndogenous
editTheNa+/K+-ATPase is upregulated bycAMP.[28]Thus, substances causing an increase in cAMP upregulate theNa+/K+-ATPase. These include the ligands of theGs-coupled GPCRs. In contrast, substances causing a decrease in cAMP downregulate theNa+/K+-ATPase. These include the ligands of theGi-coupled GPCRs. Note: Early studies indicated theoppositeeffect, but these were later found to be inaccurate due to additional complicating factors.[citation needed]
TheNa+/K+-ATPase is endogenously negatively regulated by the inositol pyrophosphate 5-InsP7, an intracellular signaling molecule generated byIP6K1,which relieves an autoinhibitory domain ofPI3Kp85αto drive endocytosis and degradation.[29]
TheNa+/K+-ATPase is also regulated by reversible phosphorylation. Research has shown that in estivating animals, theNa+/K+-ATPase is in the phosphorylated and low activity form. Dephosphorylation ofNa+/K+-ATPase can recover it to the high activity form.[13]
Exogenous
editTheNa+/K+-ATPase can be pharmacologically modified by administering drugs exogenously. Its expression can also be modified through hormones such astriiodothyronine,athyroidhormone.[13][30]
For instance,Na+/K+-ATPase found in the membrane of heart cells is an important target ofcardiac glycosides(for exampledigoxinandouabain),inotropicdrugs used to improveheartperformance by increasing its force of contraction.
Muscle contraction is dependent on a 100- to 10,000-times-higher-than-resting intracellularCa2+concentration, which is caused byCa2+release from the muscle cells' sarcoplasmic reticulum. Immediately after muscle contraction, intracellularCa2+is quickly returned to its normal concentration by a carrier enzyme in the plasma membrane, and a calcium pump insarcoplasmic reticulum,causing the muscle to relax.
According to the Blaustein-hypothesis,[31]this carrier enzyme (Na+/Ca2+exchanger, NCX) uses the Na gradient generated by theNa+-K+pump to removeCa2+from the intracellular space, hence slowing down theNa+-K+pump results in a permanently elevatedCa2+level in themuscle,which may be the mechanism of the long-term inotropic effect of cardiac glycosides such as digoxin. The problem with this hypothesis is that at pharmacological concentrations of digitalis, less than 5% of Na/K-ATPase molecules – specifically the α2 isoform in heart and arterial smooth muscle (Kd= 32 nM) – are inhibited, not enough to affect the intracellular concentration ofNa+.However, apart from the population of Na/K-ATPase in the plasma membrane, responsible for ion transport, there is another population in thecaveolaewhich acts as digitalis receptor and stimulates theEGF receptor.[32][33][34][35]
Pharmacological regulation
editIn certain conditions such as in the case of cardiac disease, theNa+/K+-ATPase may need to be inhibited via pharmacological means. A commonly used inhibitor used in the treatment of cardiac disease is digoxin (acardiac glycoside) which essentially binds "to the extracellular part of enzyme i.e. that binds potassium, when it is in a phosphorylated state, to transfer potassium inside the cell"[36]After this essential binding occurs, a dephosphorylation of the alpha subunit occurs which reduces the effect of cardiac disease. It is via the inhibiting of theNa+/K+-ATPase that sodium levels will begin to increase within the cell which ultimately increases the concentration of intracellular calcium via the sodium-calcium exchanger. This increased presence of calcium is what allows for the force of contraction to be increased. In the case of patients where the heart is not pumping hard enough to provide what is needed for the body, use of digoxin helps to temporarily overcome this.
Discovery
editNa+/K+-ATPase was proposed byJens Christian Skouin 1957 while working as assistant professor at the Department of Physiology,University of Aarhus,Denmark.He published his work that year.[37]
In 1997, he received one-half of theNobel Prize in Chemistry"for the first discovery of an ion-transporting enzyme,Na+,K+-ATPase. "[38]
Genes
edit- Alpha:ATP1A1,ATP1A2,ATP1A3,ATP1A4.ATP1A1 is expressed ubiquitously in vertebrates, and ATP1A3 in neural tissue. ATP1A2 is also known as "alpha(+)". ATP1A4 is specific to mammals.
- Beta:ATP1B1,ATP1B2,ATP1B3
ATP1B4,although closely related to ATP1B1, ATP1B2, and ATP1B3, lost its function asNa+/K+-ATPase beta subunit.[39]
The parallel evolution of resistance to cardiotonic steroids in many vertebrates
editSeveral studies have detailed the evolution of cardiotonic steroid resistance of the alpha-subunit gene family of Na/K-ATPase (ATP1A) in vertebrates via amino acid substitutions most often located in the first extracellular loop domain.[40][41][42][43][44][45][46]Amino acid substitutions conferring cardiotonic steroid resistance have evolved independently many times in all major groups of tetrapods.[44]ATP1A1 has been duplicated in some groups of frogs and neofunctionlised duplicates carry the same cardiotonic steroid resistance substitutions (Q111R and N122D) found in mice, rats and other muroids.[47][40][41][42]
In insects
editInDrosophila melanogaster,the alpha-subunit ofNa+/K+-ATPase has two paralogs, ATPα (ATPα1) and JYalpha (ATPα2), resulting from an ancient duplication in insects.[48]In Drosophila, ATPα1 is ubiquitously and highly expressed, whereas ATPα2 is most highly expressed in male testes and is essential for male fertility. Insects have at least one copy of both genes, and occasionally duplications. Low expression of ATPα2 has also been noted in other insects. Duplications andneofunctionalizationof ATPα1 have been observed in insects that are adapted to cardiotonic steroid toxins such ascardenolidesandbufadienolides.[48][49][50]Insects adapted to cardiotonic steroids typically have a number of amino acid substitutions, most often in the first extra-cellular loop of ATPα1, that confer resistance to cardiotonic steroid inhibition.[51][52]
See also
editReferences
edit- ^abGagnon KB, Delpire E (2021)."Sodium Transporters in Human Health and Disease (Figure 2)".Frontiers in Physiology.11:588664.doi:10.3389/fphys.2020.588664.PMC7947867.PMID33716756.
- ^Clausen MV, Hilbers F, Poulsen H (June 2017)."The Structure and Function of the Na,K-ATPase Isoforms in Health and Disease".Frontiers in Physiology.8:371.doi:10.3389/fphys.2017.00371.PMC5459889.PMID28634454.
- ^abHall JE, Guyton AC (2006).Textbook of medical physiology.St. Louis, Mo: Elsevier Saunders.ISBN978-0-7216-0240-0.
- ^Voet D, Voet JG (December 2010). "Section 20-3: ATP-Driven Active Transport".Biochemistry(4th ed.). John Wiley & Sons. p. 759.ISBN978-0-470-57095-1.
- ^Howarth C, Gleeson P, Attwell D (July 2012)."Updated energy budgets for neural computation in the neocortex and cerebellum".Journal of Cerebral Blood Flow and Metabolism.32(7): 1222–32.doi:10.1038/jcbfm.2012.35.PMC3390818.PMID22434069.
- ^Sanders MJ, Simon LM, Misfeldt DS (March 1983). "Transepithelial transport in cell culture: bioenergetics of Na-, D-glucose-coupled transport".Journal of Cellular Physiology.114(3): 263–6.doi:10.1002/jcp.1041140303.PMID6833401.S2CID22543559.
- ^Lynch RM, Paul RJ (March 1987). "Compartmentation of carbohydrate metabolism in vascular smooth muscle".The American Journal of Physiology.252(3 Pt 1): C328-34.doi:10.1152/ajpcell.1987.252.3.c328.PMID3030131.
- ^Glitsch HG, Tappe A (January 1993). "The Na+/K+pump of cardiac Purkinje cells is preferentially fuelled by glycolytic ATP production ".Pflügers Archiv.422(4): 380–5.doi:10.1007/bf00374294.PMID8382364.S2CID25076348.
- ^Dutka TL, Lamb GD (September 2007). "Na+-K+pumps in the transverse tubular system of skeletal muscle fibers preferentially use ATP from glycolysis ".American Journal of Physiology. Cell Physiology.293(3): C967-77.doi:10.1152/ajpcell.00132.2007.PMID17553934.S2CID2291836.
- ^Watanabe D, Wada M (December 2019). "Effects of reduced muscle glycogen on excitation-contraction coupling in rat fast-twitch muscle: a glycogen removal study".Journal of Muscle Research and Cell Motility.40(3–4): 353–364.doi:10.1007/s10974-019-09524-y.PMID31236763.S2CID195329741.
- ^Jensen R, Nielsen J, Ørtenblad N (February 2020)."Inhibition of glycogenolysis prolongs action potential repriming period and impairs muscle function in rat skeletal muscle".The Journal of Physiology.598(4): 789–803.doi:10.1113/JP278543.PMID31823376.S2CID209317559.
- ^Armstrong CM (May 2003)."The Na/K pump, Cl ion, and osmotic stabilization of cells".Proceedings of the National Academy of Sciences of the United States of America.100(10): 6257–62.Bibcode:2003PNAS..100.6257A.doi:10.1073/pnas.0931278100.PMC156359.PMID12730376.
- ^abcRamnanan CJ, Storey KB (February 2006)."Suppression of Na+/K+-ATPase activity during estivation in the land snailOtala lactea".The Journal of Experimental Biology.209(Pt 4): 677–88.doi:10.1242/jeb.02052.PMID16449562.S2CID39271006.
- ^Yuan Z, Cai T, Tian J, Ivanov AV, Giovannucci DR, Xie Z (September 2005)."Na/K-ATPase tethers phospholipase C and IP3 receptor into a calcium-regulatory complex".Molecular Biology of the Cell.16(9): 4034–45.doi:10.1091/mbc.E05-04-0295.PMC1196317.PMID15975899.
- ^Tian J, Cai T, Yuan Z, Wang H, Liu L, Haas M, et al. (January 2006)."Binding of Src to Na+/K+-ATPase forms a functional signaling complex ".Molecular Biology of the Cell.17(1): 317–26.doi:10.1091/mbc.E05-08-0735.PMC1345669.PMID16267270.
- ^Li Z, Cai T, Tian J, Xie JX, Zhao X, Liu L, et al. (July 2009)."NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells".The Journal of Biological Chemistry.284(31): 21066–76.doi:10.1074/jbc.M109.013821.PMC2742871.PMID19506077.
- ^Lee K, Jung J, Kim M, Guidotti G (January 2001)."Interaction of the alpha subunit of Na,K-ATPase with cofilin".The Biochemical Journal.353(Pt 2): 377–85.doi:10.1042/0264-6021:3530377.PMC1221581.PMID11139403.
- ^Forrest MD, Wall MJ, Press DA, Feng J (December 2012)."The sodium-potassium pump controls the intrinsic firing of the cerebellar Purkinje neuron".PLOS ONE.7(12): e51169.Bibcode:2012PLoSO...751169F.doi:10.1371/journal.pone.0051169.PMC3527461.PMID23284664.
- ^Zylbertal A, Kahan A, Ben-Shaul Y, Yarom Y, Wagner S (December 2015)."Prolonged Intracellular Na+Dynamics Govern Electrical Activity in Accessory Olfactory Bulb Mitral Cells ".PLOS Biology.13(12): e1002319.doi:10.1371/journal.pbio.1002319.PMC4684409.PMID26674618.
- ^Zylbertal A, Yarom Y, Wagner S (2017)."The Slow Dynamics of Intracellular Sodium Concentration Increase the Time Window of Neuronal Integration: A Simulation Study".Frontiers in Computational Neuroscience.11:85.doi:10.3389/fncom.2017.00085.PMC5609115.PMID28970791.
- ^Forrest MD (December 2014)."The sodium-potassium pump is an information processing element in brain computation".Frontiers in Physiology.5(472): 472.doi:10.3389/fphys.2014.00472.PMC4274886.PMID25566080.
- ^Cannon SC (July 2004)."Paying the price at the pump: dystonia from mutations in a Na+/K+-ATPase ".Neuron.43(2): 153–4.doi:10.1016/j.neuron.2004.07.002.PMID15260948.
- ^Calderon DP, Fremont R, Kraenzlin F, Khodakhah K (March 2011)."The neural substrates of rapid-onset Dystonia-Parkinsonism".Nature Neuroscience.14(3): 357–65.doi:10.1038/nn.2753.PMC3430603.PMID21297628.
- ^Forrest MD (April 2015)."Simulation of alcohol action upon a detailed Purkinje neuron model and a simpler surrogate model that runs >400 times faster".BMC Neuroscience.16(27): 27.doi:10.1186/s12868-015-0162-6.PMC4417229.PMID25928094.
- ^Forrest M (4 April 2015)."The Neuroscience Reason We Fall Over When Drunk".Science 2.0.Retrieved30 May2018.
- ^Young EA, Fowler CD, Kidd GJ, Chang A, Rudick R, Fisher E, Trapp BD (April 2008). "Imaging correlates of decreased axonal Na+/K+ATPase in chronic multiple sclerosis lesions ".Annals of Neurology.63(4): 428–35.doi:10.1002/ana.21381.PMID18438950.S2CID14658965.
- ^Smith RS, Florio M, Akula SK, Neil JE, Wang Y, Hill RS, et al. (June 2021)."Early role for a Na+,K+-ATPase (ATP1A3) in brain development ".Proceedings of the National Academy of Sciences of the United States of America.118(25): e2023333118.Bibcode:2021PNAS..11823333S.doi:10.1073/pnas.2023333118.PMC8237684.PMID34161264.
- ^Burnier M (2008).Sodium In Health And Disease.CRC Press. p. 15.ISBN978-0-8493-3978-3.
- ^Chin AC, Gao Z, Riley AM, Furkert D, Wittwer C, Dutta A, et al. (October 2020)."The inositol pyrophosphate 5-InsP7drives sodium-potassium pump degradation by relieving an autoinhibitory domain of PI3K p85α ".Science Advances.6(44): eabb8542.Bibcode:2020SciA....6.8542C.doi:10.1126/sciadv.abb8542.PMC7608788.PMID33115740.S2CID226036261.
- ^Lin HH, Tang MJ (January 1997). "Thyroid hormone upregulates Na,K-ATPase α and β mRNA in primary cultures of proximal tubule cells".Life Sciences.60(6): 375–382.doi:10.1016/S0024-3205(96)00661-3.PMID9031683.
- ^Blaustein MP (May 1977). "Sodium ions, calcium ions, blood pressure regulation, and hypertension: a reassessment and a hypothesis".The American Journal of Physiology.232(5): C165-73.doi:10.1152/ajpcell.1977.232.5.C165.PMID324293.S2CID9814212.
- ^Schoner W, Scheiner-Bobis G (September 2008). "Role of endogenous cardiotonic steroids in sodium homeostasis".Nephrology, Dialysis, Transplantation.23(9): 2723–9.doi:10.1093/ndt/gfn325.PMID18556748.
- ^Blaustein MP, Hamlyn JM (December 2010)."Signaling mechanisms that link salt retention to hypertension: endogenous ouabain, the Na+pump, the Na+/Ca2+exchanger and TRPC proteins ".Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease.1802(12): 1219–29.doi:10.1016/j.bbadis.2010.02.011.PMC2909369.PMID20211726.
- ^Fuerstenwerth H (2014). "On the differences between ouabain and digitalis glycosides".American Journal of Therapeutics.21(1): 35–42.doi:10.1097/MJT.0b013e318217a609.PMID21642827.S2CID20180376.
- ^Pavlovic D (2014). "The role of cardiotonic steroids in the pathogenesis of cardiomyopathy in chronic kidney disease".Nephron Clinical Practice.128(1–2): 11–21.doi:10.1159/000363301.PMID25341357.S2CID2066801.
- ^"Na+/K+-ATPase and inhibitors (Digoxin) ".Pharmacorama.Archived fromthe originalon 2020-09-28.Retrieved2019-11-08.
- ^Skou JC (February 1957). "The influence of some cations on an adenosine triphosphatase from peripheral nerves".Biochimica et Biophysica Acta.23(2): 394–401.doi:10.1016/0006-3002(57)90343-8.PMID13412736.S2CID32516710.
- ^"The Nobel Prize in Chemistry 1997".NobelPrize.org.Nobel Media AB. 15 October 1997.
- ^"ATPase Na+/K+ transporting subunits (ATP1)".HGNC.Retrieved26 June2024.
- ^abMoore, David J.; Halliday, Damien C. T.; Rowell, David M.; Robinson, Anthony J.; Keogh, J. Scott (2009-08-23)."Positive Darwinian selection results in resistance to cardioactive toxins in true toads (Anura: Bufonidae)".Biology Letters.5(4): 513–516.doi:10.1098/rsbl.2009.0281.ISSN1744-9561.PMC2781935.PMID19465576.
- ^abHernández Poveda M (2022) Convergent evolution of neo-functionalized duplications of ATP1A1 in dendrobatid and grass frogs. MS Thesis Dissertation. Universidad de los Andes
- ^abMohammadi, Shabnam; Yang, Lu; Harpak, Arbel; Herrera-Álvarez, Santiago; Rodríguez-Ordoñez, María del Pilar; Peng, Julie; Zhang, Karen; Storz, Jay F.; Dobler, Susanne; Crawford, Andrew J.; Andolfatto, Peter (2021-06-21)."Concerted evolution reveals co-adapted amino acid substitutions in frogs that prey on toxic toads".Current Biology.31(12): 2530–2538.e10.doi:10.1016/j.cub.2021.03.089.ISSN0960-9822.PMC8281379.PMID33887183.
- ^Mohammadi, Shabnam; Brodie, Edmund D.; Neuman-Lee, Lorin A.; Savitzky, Alan H. (2016-05-01)."Mutations to the cardiotonic steroid binding site of Na+/K+-ATPase are associated with high level of resistance to gamabufotalin in a natricine snake".Toxicon.114:13–15.doi:10.1016/j.toxicon.2016.02.019.ISSN0041-0101.PMID26905927.
- ^abMohammadi, Shabnam; Herrera-Álvarez, Santiago; Yang, Lu; Rodríguez-Ordoñez, María del Pilar; Zhang, Karen; Storz, Jay F.; Dobler, Susanne; Crawford, Andrew J.; Andolfatto, Peter (2022-08-16)."Constraints on the evolution of toxin-resistant Na,K-ATPases have limited dependence on sequence divergence".PLOS Genetics.18(8): e1010323.doi:10.1371/journal.pgen.1010323.ISSN1553-7390.PMC9462791.PMID35972957.
- ^Mohammadi, Shabnam; Özdemir, Halil İbrahim; Ozbek, Pemra; Sumbul, Fidan; Stiller, Josefin; Deng, Yuan; Crawford, Andrew J; Rowland, Hannah M; Storz, Jay F; Andolfatto, Peter; Dobler, Susanne (2022-12-06)."Epistatic Effects Between Amino Acid Insertions and Substitutions Mediate Toxin resistance of Vertebrate Na+,K+-ATPases".Molecular Biology and Evolution.39(12): msac258.doi:10.1093/molbev/msac258.ISSN0737-4038.PMC9778839.PMID36472530.
- ^Ujvari, Beata; Mun, Hee-chang; Conigrave, Arthur D.; Bray, Alessandra; Osterkamp, Jens; Halling, Petter; Madsen, Thomas (January 2013)."Isolation Breeds Naivety: Island Living Robs Australian Varanid Lizards of Toad-Toxin Immunity Via Four-Base-Pair Mutation".Evolution.67(1): 289–294.doi:10.1111/j.1558-5646.2012.01751.x.PMID23289579.
- ^Price, Elmer M.; Lingrel, Jerry B. (1988-11-01)."Structure-function relationships in the sodium-potassium ATPase.alpha. subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme".Biochemistry.27(22): 8400–8408.doi:10.1021/bi00422a016.ISSN0006-2960.PMID2853965.
- ^abZhen, Ying; Aardema, Matthew L.; Medina, Edgar M.; Schumer, Molly; Andolfatto, Peter (2012-09-28)."Parallel Molecular Evolution in an Herbivore Community".Science.337(6102): 1634–1637.Bibcode:2012Sci...337.1634Z.doi:10.1126/science.1226630.ISSN0036-8075.PMC3770729.PMID23019645.
- ^Yang, L.; Ravikanthachari, N.; Mariño-Pérez, R.; Deshmukh, R.; Wu, M.; Rosenstein, A.; Kunte, K.; Song, H.; Andolfatto, P. (2019)."Predictability in the evolution of Orthopteran cardenolide insensitivity".Philosophical Transactions of the Royal Society of London, Series B.374(1777): 20180246.doi:10.1098/rstb.2018.0246.PMC6560278.PMID31154978.
- ^Petschenka Georg, Vera Wagschal, Michael von Tschirnhaus, Alexander Donath, Susanne Dobler 2017Petschenka, G.; Wagschal, V.; von Tschirnhaus, M.; Donath, A.; Dobler, S. (2017)."Convergently Evolved Toxic Secondary Metabolites in Plants Drive the Parallel Molecular Evolution of Insect Resistance".The American Naturalist.190(S1): S29–S43.doi:10.1086/691711.PMID28731826.S2CID3908073.
- ^Labeyrie E, Dobler S (2004)."Molecular adaptation of Chrysochus leaf beetles to toxic compounds in their food plants".Molecular Biology and Evolution.21(2): 218–21.doi:10.1093/molbev/msg240.PMID12949136.
- ^Dobler, Susanne; Dalla, Safaa; Wagschal, Vera; Agrawal, Anurag A. (2012)."Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase".Proceedings of the National Academy of Sciences.109(32): 13040–13045.doi:10.1073/pnas.1202111109.PMC3420205.PMID22826239.
External links
edit- Sodium,+Potassium+ATPaseat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
- RCSB Protein Data Bank: Sodium–Potassium Pump
- A videobyKhan Academy.