TheThermoproteotaareprokaryotesthat have been classified as aphylumof the domainArchaea.[2][3][4]Initially, the Thermoproteota were thought to be sulfur-dependentextremophilesbut recent studies have identified characteristic Thermoproteota environmentalrRNAindicating the organisms may be the most abundant archaea in the marine environment.[5]Originally, they were separated from the other archaea based on rRNA sequences; other physiological features, such as lack ofhistones,have supported this division, although some crenarchaea were found to have histones.[6]Until 2005 all cultured Thermoproteota had been thermophilic or hyperthermophilic organisms, some of which have the ability to grow at up to 113 °C.[7]These organisms stainGram negativeand are morphologically diverse, having rod,cocci,filamentousand oddly-shaped cells.[8]Recent evidence shows that some members of the Thermoproteota are methanogens.
| Thermoproteota | |
|---|---|
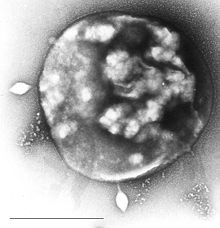
| |
| ArchaeaSulfolobusinfected with specific virusSTSV-1. | |
| Scientific classification | |
| Domain: | Archaea |
| Kingdom: | Proteoarchaeota |
| Superphylum: | TACK group |
| Phylum: | Thermoproteota Garrity & Holt 2021[1] |
| Class | |
| |
| Synonyms | |
| |
Thermoproteota were initially classified as a part ofRegnumEocytain 1984,[9]but this classification has been discarded. The term "eocyte" now applies to eitherTACK(formerly Crenarchaeota) or to Thermoproteota.
Sulfolobus
editOne of the best characterized members of the Crenarchaeota isSulfolobus solfataricus.This organism was originally isolated fromgeothermally heatedsulfuric springs in Italy, and grows at 80 °C and pH of 2–4.[10]Since its initial characterization byWolfram Zillig,a pioneer in thermophile and archaean research, similar species in the samegenushave been found around the world. Unlike the vast majority of cultured thermophiles,Sulfolobusgrowsaerobicallyandchemoorganotrophically(gaining its energy from organic sources such as sugars). These factors allow a much easier growth under laboratory conditions thananaerobic organismsand have led toSulfolobusbecoming a model organism for the study of hyperthermophiles and a large group of diverse viruses that replicate within them.
| 16S rRNA basedLTP_06_2022[11][12][13] | 53 marker proteins basedGTDB08-RS214[14][15][16] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Recombinational repair of DNA damage
editIrradiation ofS. solfataricuscells withultravioletlight strongly induces formation oftype IV pilithat can then promote cellular aggregation.[17]Ultraviolet light-induced cellular aggregation was shown by Ajon et al.[18]to mediate high frequency inter-cellularchromosomemarker exchange. Cultures that were ultraviolet light-induced had recombination rates exceeding those of uninduced cultures by as much as three orders of magnitude.S. solfataricuscells are only able to aggregate with other members of their own species.[18]Frols et al.[17][19]and Ajon et al.[18]considered that the ultraviolet light-inducible DNA transfer process, followed byhomologous recombinationalrepair ofdamaged DNA,is an important mechanism for promoting chromosome integrity.
This DNA transfer process can be regarded as a primitive form ofsexual interaction.
Marine species
editBeginning in 1992, data were published that reported sequences of genes belonging to the Thermoproteota in marine environments.[20],[21]Since then, analysis of the abundantlipidsfrom the membranes of Thermoproteota taken from the open ocean have been used to determine the concentration of these “low temperature Crenarchaea” (SeeTEX-86). Based on these measurements of their signature lipids, Thermoproteota are thought to be very abundant and one of the main contributors to thefixation of carbon.[citation needed]DNA sequences from Thermoproteota have also been found in soil and freshwater environments, suggesting that this phylum is ubiquitous to most environments.[22]
In 2005, evidence of the first cultured “low temperature Crenarchaea” was published. NamedNitrosopumilus maritimus,it is anammonia-oxidizing organism isolated from a marine aquarium tank and grown at 28 °C.[23]
Possible connections with eukaryotes
editThe research abouttwo-domain systemof classification has paved the possibilities of connections between crenarchaea andeukaryotes.[24]
DNA analysis from 2008 (and later, 2017) has shown that eukaryotes possible evolved from thermoproteota-like organisms. Other candidates for the ancestor of eukaryotes include closely relatedasgards.This could suggest that eukaryotic organisms possibly evolved from prokaryotes.
These results are similar to theeocyte hypothesisof 1984, proposed byJames A. Lake.[9]The classification according to Lake, states that both crenarchaea and asgards belong to Kingdom Eocyta. Though this has been discarded by scientists, the main concept remains. The term "Eocyta" now either refers to theTACK groupor to Phylum Thermoproteota itself.
However, the topic is highly debated and research is still going on.
See also
editReferences
edit- ^Oren A, Garrity GM (2021)."Valid publication of the names of forty-two phyla of prokaryotes".Int J Syst Evol Microbiol.71(10): 5056.doi:10.1099/ijsem.0.005056.PMID34694987.S2CID239887308.
- ^See theNCBIwebpage on Crenarchaeota
- ^C.Michael Hogan. 2010.Archaea.eds. E.Monosson & C.Cleveland, Encyclopedia of Earth. National Council for Science and the Environment, Washington DC.
- ^Data extracted from the"NCBI taxonomy resources".National Center for Biotechnology Information.Retrieved2007-03-19.
- ^Madigan M; Martinko J, eds. (2005).Brock Biology of Microorganisms(11th ed.). Prentice Hall.ISBN978-0-13-144329-7.
- ^Cubonova L, Sandman K, Hallam SJ, Delong EF, Reeve JN (2005)."Histones in Crenarchaea".Journal of Bacteriology.187(15): 5482–5485.doi:10.1128/JB.187.15.5482-5485.2005.PMC1196040.PMID16030242.
- ^Blochl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch HW, Stetter KO (1997). "Pyrolobus fumarii,gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 °C ".Extremophiles.1(1): 14–21.doi:10.1007/s007920050010.PMID9680332.S2CID29789667.
- ^Garrity GM, Boone DR, eds. (2001).Bergey's Manual of Systematic Bacteriology Volume 1: The Archaea and the Deeply Branching and Phototrophic Bacteria(2nd ed.). Springer.ISBN978-0-387-98771-2.
- ^abLake JA, Henderson E, Oakes M, Clark MW (June 1984)."Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes".Proceedings of the National Academy of Sciences of the United States of America.81(12): 3786–3790.Bibcode:1984PNAS...81.3786L.doi:10.1073/pnas.81.12.3786.PMC345305.PMID6587394.
- ^Zillig W, Stetter KO, Wunderl S, Schulz W, Priess H, Scholz I (1980). "The Sulfolobus-" Caldariellard "group: Taxonomy on the basis of the structure of DNA-dependent RNA polymerases".Arch. Microbiol.125(3): 259–269.Bibcode:1980ArMic.125..259Z.doi:10.1007/BF00446886.S2CID5805400.
- ^"The LTP".Retrieved10 May2023.
- ^"LTP_all tree in newick format".Retrieved10 May2023.
- ^"LTP_06_2022 Release Notes"(PDF).Retrieved10 May2023.
- ^"GTDB release 08-RS214".Genome Taxonomy Database.Retrieved10 May2023.
- ^"ar53_r214.sp_label".Genome Taxonomy Database.Retrieved10 May2023.
- ^"Taxon History".Genome Taxonomy Database.Retrieved10 May2023.
- ^abFröls S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, Boekema EJ, Driessen AJ, Schleper C, Albers SV. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol. 2008 Nov;70(4):938-52. doi: 10.1111/j.1365-2958.2008.06459.x. PMID 18990182
- ^abcAjon M, Fröls S, van Wolferen M, Stoecker K, Teichmann D, Driessen AJ, Grogan DW, Albers SV, Schleper C. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol Microbiol. 2011 Nov;82(4):807-17. doi: 10.1111/j.1365-2958.2011.07861.x. Epub 2011 Oct 18. PMID 21999488
- ^Fröls S, White MF, Schleper C. Reactions to UV damage in the model archaeon Sulfolobus solfataricus. Biochem Soc Trans. 2009 Feb;37(Pt 1):36-41. doi: 10.1042/BST0370036. PMID 19143598
- ^Fuhrman JA, McCallum K, Davis AA (1992). "Novel major archaebacterial group from marine plankton".Nature.356(6365): 148–9.Bibcode:1992Natur.356..148F.doi:10.1038/356148a0.PMID1545865.S2CID4342208.
- ^DeLong EF (1992)."Archaea in coastal marine environments".Proc Natl Acad Sci USA.89(12): 5685–9.Bibcode:1992PNAS...89.5685D.doi:10.1073/pnas.89.12.5685.PMC49357.PMID1608980.
- ^Barns SM, Delwiche CF, Palmer JD, Pace NR (1996)."Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences".Proc Natl Acad Sci USA.93(17): 9188–93.Bibcode:1996PNAS...93.9188B.doi:10.1073/pnas.93.17.9188.PMC38617.PMID8799176.
- ^Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005). "Isolation of an autotrophic ammonia-oxidizing marine archaeon".Nature.437(7058): 543–6.Bibcode:2005Natur.437..543K.doi:10.1038/nature03911.PMID16177789.S2CID4340386.
- ^Yutin, Natalya; Makarova, Kira S.; Mekhedov, Sergey L.; Wolf, Yuri I.; Koonin, Eugene V. (2008)."The deep archaeal roots of eukaryotes".Molecular Biology and Evolution.25(8): 1619–1630.doi:10.1093/molbev/msn108.PMC2464739.PMID18463089.
Further reading
editScientific journals
edit- Cavalier-Smith, T (2002)."The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification".Int. J. Syst. Evol. Microbiol.52(Pt 1): 7–76.doi:10.1099/00207713-52-1-7.PMID11837318.
- Stackebrandt, E; Frederiksen W; Garrity GM; Grimont PA; Kampfer P; Maiden MC; Nesme X; Rossello-Mora R; Swings J; Truper HG; Vauterin L; Ward AC; Whitman WB (2002). "Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology".Int. J. Syst. Evol. Microbiol.52(Pt 3): 1043–1047.doi:10.1099/00207713-52-3-1043.PMID12054223.
- Gurtler, V; Mayall BC (2001). "Genomic approaches to typing, taxonomy and evolution of bacterial isolates".Int. J. Syst. Evol. Microbiol.51(Pt 1): 3–16.doi:10.1099/00207713-51-1-3.PMID11211268.
- Dalevi, D; Hugenholtz P; Blackall LL (2001)."A multiple-outgroup approach to resolving division-level phylogenetic relationships using 16S rDNA data".Int. J. Syst. Evol. Microbiol.51(Pt 2): 385–391.doi:10.1099/00207713-51-2-385.PMID11321083.
- Keswani, J; Whitman WB (2001)."Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes".Int. J. Syst. Evol. Microbiol.51(Pt 2): 667–678.doi:10.1099/00207713-51-2-667.PMID11321113.
- Young, JM (2001). "Implications of alternative classifications and horizontal gene transfer for bacterial taxonomy".Int. J. Syst. Evol. Microbiol.51(Pt 3): 945–953.doi:10.1099/00207713-51-3-945.PMID11411719.
- Christensen, H; Bisgaard M; Frederiksen W; Mutters R; Kuhnert P; Olsen JE (2001)."Is characterization of a single isolate sufficient for valid publication of a new genus or species? Proposal to modify recommendation 30b of the Bacteriological Code (1990 Revision)".Int. J. Syst. Evol. Microbiol.51(Pt 6): 2221–2225.doi:10.1099/00207713-51-6-2221.PMID11760965.
- Christensen, H; Angen O; Mutters R; Olsen JE; Bisgaard M (2000)."DNA-DNA hybridization determined in micro-wells using covalent attachment of DNA".Int. J. Syst. Evol. Microbiol.50(3): 1095–1102.doi:10.1099/00207713-50-3-1095.PMID10843050.
- Xu, HX; Kawamura Y; Li N; Zhao L; Li TM; Li ZY; Shu S; Ezaki T (2000)."A rapid method for determining the G+C content of bacterial chromosomes by monitoring fluorescence intensity during DNA denaturation in a capillary tube".Int. J. Syst. Evol. Microbiol.50(4): 1463–1469.doi:10.1099/00207713-50-4-1463.PMID10939651.
- Young, JM (2000)."Suggestions for avoiding on-going confusion from the Bacteriological Code".Int. J. Syst. Evol. Microbiol.50(4): 1687–1689.doi:10.1099/00207713-50-4-1687.PMID10939677.
- Hansmann, S; Martin W (2000)."Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis".Int. J. Syst. Evol. Microbiol.50(4): 1655–1663.doi:10.1099/00207713-50-4-1655.PMID10939673.
- Tindall, BJ (1999)."Proposal to change the Rule governing the designation of type strains deposited under culture collection numbers allocated for patent purposes".Int. J. Syst. Bacteriol.49(3): 1317–1319.doi:10.1099/00207713-49-3-1317.PMID10490293.
- Tindall, BJ (1999)."Proposal to change Rule 18a, Rule 18f and Rule 30 to limit the retroactive consequences of changes accepted by the ICSB".Int. J. Syst. Bacteriol.49(3): 1321–1322.doi:10.1099/00207713-49-3-1321.PMID10425797.
- Tindall, BJ (1999)."Misunderstanding the Bacteriological Code".Int. J. Syst. Bacteriol.49(3): 1313–1316.doi:10.1099/00207713-49-3-1313.PMID10425796.
- Tindall, BJ (1999)."Proposals to update and make changes to the Bacteriological Code".Int. J. Syst. Bacteriol.49(3): 1309–1312.doi:10.1099/00207713-49-3-1309.PMID10425795.
- Burggraf, S; Huber H; Stetter KO (1997)."Reclassification of the crenarchael orders and families in accordance with 16S rRNA sequence data".Int. J. Syst. Bacteriol.47(3): 657–660.doi:10.1099/00207713-47-3-657.PMID9226896.
- Palys, T; Nakamura LK; Cohan FM (1997)."Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data".Int. J. Syst. Bacteriol.47(4): 1145–1156.doi:10.1099/00207713-47-4-1145.PMID9336922.
- Euzeby, JP (1997)."List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet".Int. J. Syst. Bacteriol.47(2): 590–592.doi:10.1099/00207713-47-2-590.PMID9103655.
- Clayton, RA; Sutton G; Hinkle PS Jr; Bult C; Fields C (1995)."Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa".Int. J. Syst. Bacteriol.45(3): 595–599.doi:10.1099/00207713-45-3-595.PMID8590690.
- Murray, RG; Schleifer KH (1994)."Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes".Int. J. Syst. Bacteriol.44(1): 174–176.doi:10.1099/00207713-44-1-174.PMID8123559.
- Winker, S; Woese CR (1991). "A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics".Syst. Appl. Microbiol.14(4): 305–310.Bibcode:1991SyApM..14..305W.doi:10.1016/s0723-2020(11)80303-6.PMID11540071.
- Woese, CR; Kandler O; Wheelis ML (1990)."Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya".Proc. Natl. Acad. Sci. USA.87(12): 4576–4579.Bibcode:1990PNAS...87.4576W.doi:10.1073/pnas.87.12.4576.PMC54159.PMID2112744.
- Achenbach-Richter, L; Woese CR (1988). "The ribosomal gene spacer region in archaebacteria".Syst. Appl. Microbiol.10(3): 211–214.Bibcode:1988SyApM..10..211A.doi:10.1016/s0723-2020(88)80002-x.PMID11542149.
- McGill, TJ; Jurka J; Sobieski JM; Pickett MH; Woese CR; Fox GE (1986). "Characteristic archaebacterial 16S rRNA oligonucleotides".Syst. Appl. Microbiol.7(2–3): 194–197.Bibcode:1986SyApM...7..194M.doi:10.1016/S0723-2020(86)80005-4.PMID11542064.
- Woese, CR; Gupta R; Hahn CM; Zillig W; Tu J (1984). "The phylogenetic relationships of three sulfur dependent archaebacteria".Syst. Appl. Microbiol.5(1): 97–105.Bibcode:1984SyApM...5...97W.doi:10.1016/S0723-2020(84)80054-5.PMID11541975.
- Woese, CR; Olsen GJ (1984). "The phylogenetic relationships of three sulfur dependent archaebacteria".Syst. Appl. Microbiol.5:97–105.Bibcode:1984SyApM...5...97W.doi:10.1016/S0723-2020(84)80054-5.PMID11541975.
- Woese, CR; Fox GE (1977)."Phylogenetic structure of the prokaryotic domain: the primary kingdoms".Proc. Natl. Acad. Sci. USA.74(11): 5088–5090.Bibcode:1977PNAS...74.5088W.doi:10.1073/pnas.74.11.5088.PMC432104.PMID270744.
Scientific handbooks
edit- Garrity GM, Holt JG (2001)."Phylum AI. Crenarchaeota phy. nov.".In DR Boone, RW Castenholz (eds.).Bergey's Manual of Systematic Bacteriology Volume 1: The Archaea and the deeply branching and phototrophic Bacteria(2nd ed.). New York: Springer Verlag. pp.169.ISBN978-0-387-98771-2.
External links
edit- "Crenarchaeota".Virtual Microbiology (bact.wisc.edu).University of Wisconsin.
- "Comparative analysis of crenarchaeal genomes".Integrated Microbial Genomes System.United States Department of Energy.Archived fromthe originalon 2010-09-01.