Thecryptomonads(orcryptophytes)[2]are a group ofalgae,[3]most of which haveplastids.They are traditionally considered adivisionofalgaeamongphycologists,under the name ofCryptophyta.[4]They are common in freshwater, and also occur in marine and brackish habitats. Each cell is around10–50 μmin size and flattened in shape, with an anterior groove or pocket. At the edge of the pocket there are typically two slightly unequalflagella.Some may exhibitmixotrophy.[5]They are classified ascladeCryptomonada,which is divided into two classes: heterotrophicGoniomonadeaand phototrophicCryptophyceae.The two groups are united under three shared morphological characteristics: presence of aperiplast,ejectisomeswith secondary scroll, and mitochondrialcristaewith flat tubules.[6]Genetic studies as early as 1994 also supported the hypothesis thatGoniomonaswas sister to Cryptophyceae.[7]A study in 2018 found strong evidence that the common ancestor ofCryptomonadawas an autotrophic protist.[8]
| Cryptomonads | |
|---|---|
| |
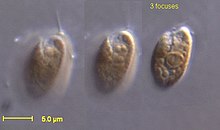
| Rhodomonas salina | |
| Scientific classification | |
| Domain: | Eukaryota |
| Clade: | Diaphoretickes |
| Clade: | CAM |
| Clade: | Pancryptista |
| Phylum: | Cryptista |
| Subphylum: | Rollomonadia |
| Superclass: | Cryptomonada Cavalier-Smith,2004 stat. nov. 2015[1] |
| Classes & orders[1] | |
Characteristics
editCryptomonads are distinguished by the presence of characteristicextrusomescalledejectosomes,which consist of two connected spiral ribbons held under tension.[9]If the cells are irritated either by mechanical, chemical or light stress, they discharge, propelling the cell in a zig-zag course away from the disturbance. Large ejectosomes, visible under the light microscope, are associated with the pocket; smaller ones occur underneath theperiplast,the cryptophyte-specific cell surrounding.[10][11]
Except for the classGoniomonadea,which lacks plastids entirely,[12]andCryptomonas paramecium(previously calledChilomonasparamecium), which hasleucoplasts,cryptomonads have one or two chloroplasts. These containchlorophyllsaandc,together withphycobiliproteinsand other pigments, and vary in color (brown, red to blueish-green). Each is surrounded by four membranes, and there is a reducedcell nucleuscalled anucleomorphbetween the middle two. This indicates that the plastid was derived from aeukaryoticsymbiont, shown by genetic studies to have been ared alga.[13]However, the plastids are very different from red algal plastids: phycobiliproteins are present but only in the thylakoid lumen and are present only as phycoerythrin orphycocyanin.In the case ofRhodomonas,the crystal structure has been determined to 1.63 Å;[14]and it has been shown that the Alpha subunit bears no relation to any other known phycobiliprotein.
A few cryptomonads, such asCryptomonas,can formpalmelloidstages, but readily escape the surrounding mucus to become free-living flagellates again. SomeCryptomonasspecies may also form immotilemicrobial cysts—resting stages with rigid cell walls to survive unfavorable conditions. Cryptomonad flagella are inserted parallel to one another, and are covered by bipartite hairs calledmastigonemes,formed within theendoplasmic reticulumand transported to the cell surface. Small scales may also be present on the flagella and cell body. Themitochondriahave flatcristae,andmitosisis open;sexual reproductionhas also been reported.
- Anterior flagellum (mastigonemeson both faces)
- Posterior flagellum (mastigonemes on one face)
- Contractile vacuole,regulates the quantity of water inside a cell
- Vestibulum
- Basal bodies
- Gullet (furrow or crypt)
- Mitochondrion,createsATP(energy) for the cell
- Maupa's bodies
- Ejectisomes
- Starch granule
- Golgi apparatus,packages proteins
- Nucleomorph,a small, vestigial eukaryotic nucleus
- Pyrenoid,center ofcarbon fixation
- Periplastidial compartment
- Thylakoid,site of thelight-dependent reactionsofphotosynthesis
- Plastidmembranes (4, secondary)
- Nucleus
- Nucleolus
- Lipid globules
Classification
editThe first mention of cryptomonads appears to have been made byChristian Gottfried Ehrenbergin 1831,[15]while studyingInfusoria.Later, botanists treated them as a separatealgaegroup, class Cryptophyceae or division Cryptophyta, while zoologists treated them as theflagellateprotozoaorder Cryptomonadina. In some classifications, the cryptomonads were considered close relatives of thedinoflagellatesbecause of their (seemingly) similar pigmentation, being grouped as thePyrrhophyta.Cryptomonad chloroplasts are closely related to those of theheterokontsandhaptophytes,and the three groups were united by Cavalier-Smith as theChromista.However, the case that the organisms themselves are closely related was counter-indicated by the major differences in cell organization (ultrastructural identity), suggesting that the three major lineages assigned to the chromists had acquired plastids independently, and that chromists are polyphyletic. The perspective that cryptomonads are primitively heterotrophic and secondarily acquired chloroplasts, is supported by molecular evidence.[16]Parfrey et al. and Burki et al. placed Cryptophyceae as a sister clade to theGreen Algae,[17]or green algae plusglaucophytes.[18]The sister group to the cryptomonads is likely the kathablepharids (also referred to as katablepharids), a group of flagellates that also have ejectisomes.[19]
One suggested grouping is as follows: (1)Cryptomonas,(2)Chroomonas/KommaandHemiselmis,(3)Rhodomonas/Rhinomonas/Storeatula,(4)Guillardia/Hanusia,(5)Geminigera/Plagioselmis/Teleaulax,(6)Proteomonas sulcata,(7)Falcomonas daucoides.[20]
References
edit- ^abCavalier-Smith, Thomas (2018)."Kingdom Chromista and its eight phyla: A new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences".Protoplasma.255:297–357.doi:10.1007/S00709-017-1147-3.PMC5756292.PMID28875267.
- ^Barnes, Richard Stephen Kent (2001).The Invertebrates: A Synthesis.Wiley-Blackwell. p. 41.ISBN978-0-632-04761-1.
- ^Khan H, Archibald JM (May 2008)."Lateral transfer of introns in the cryptophyte plastid genome".Nucleic Acids Res.36(9): 3043–53.doi:10.1093/nar/gkn095.PMC2396441.PMID18397952.
- ^Okamoto, N.; Inouye, I. (2005). "The Katablepharids are a Distant Sister Group of the Cryptophyta: A Proposal for Katablepharidophyta Divisio Nova/Kathablepharida Phylum Novum Based on SSU rDNA and Beta-Tubulin Phylogeny".Protist.156(2): 163–179.doi:10.1016/j.protis.2004.12.003.PMID16171184.
- ^"Cryptophyta - the cryptomonads".Archived fromthe originalon 2011-06-10.Retrieved2009-06-02.
- ^Cavalier-Smith, Thomas (2004).Organelles, Genomes, and Eukaryote Phylogeny.pp. 87–88.
- ^McFadden, Gilson, & Hill (1994), "Goniomonas: rRNA sequences indicate that this phagotrophic flagellate is a close relative of the host component of cryptomonads",European Journal of Phycology,29(1): 29–32,doi:10.1080/09670269400650451
{{citation}}:CS1 maint: multiple names: authors list (link) - ^Cenci (2018), "Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids",BMC Biology,16(1): 137,doi:10.1186/s12915-018-0593-5,PMC6260743,PMID30482201
- ^Graham, L. E.; Graham, J. M.; Wilcox, L. W. (2009).Algae(2nd ed.). San Francisco, CA: Benjamin Cummings (Pearson).ISBN9780321559654.
- ^Morrall, S.; Greenwood, A. D. (1980). "A comparison of the periodic sub-structures of the trichocysts of the Cryptophyceae and Prasinophyceae".BioSystems.12(1–2): 71–83.doi:10.1016/0303-2647(80)90039-8.PMID6155157.
- ^Grim, J. N.;Staehelin, L. A.(1984). "The ejectisomes of the flagellateChilomonas paramecium- Visualization by freeze-fracture and isolation techniques ".Journal of Protozoology.31(2): 259–267.doi:10.1111/j.1550-7408.1984.tb02957.x.PMID6470985.
- ^Nuclear genome sequence of the plastid-lacking cryptomonadGoniomonas avonleaprovides insights into the evolution of secondary plastids
- ^Douglas, S.; et al. (2002)."The highly reduced genome of an enslaved algal nucleus".Nature.410(6832): 1091–1096.Bibcode:2001Natur.410.1091D.doi:10.1038/35074092.PMID11323671.
- ^Wilk, K.; et al. (1999)."Evolution of a light-harvesting protein by addition of new subunits and rearrangement of conserved elements: Crystal structure of a cryptophyte phycoerythrin at 1.63Å resolution".PNAS.96(16): 8901–8906.doi:10.1073/pnas.96.16.8901.PMC17705.PMID10430868.
- ^Novarino, G. (2012)."Cryptomonad taxonomy in the 21st century: The first 200 years".Phycological Reports: Current Advances in Algal Taxonomy and Its Applications: Phylogenetic, Ecological and Applied Perspective:19–52.Retrieved2018-10-16.
- ^Cenci, U.; Sibbald, S. J.; Curtis, B. A.; Kamikawa, R.; Eme, L.; Moog, D.; Henrissat, B.; Maréchal, E.; Chabi, M.; Djemiel, C.; Roger, A. J.; Kim, E.; Archibald, J. M. (2018)."Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids".BMC Biology.16(1): 137.doi:10.1186/s12915-018-0593-5.PMC6260743.PMID30482201.
- ^Parfrey, Laura Wegener;Lahr, Daniel J. G.; Knoll, Andrew H.;Katz, Laura A.(August 16, 2011)."Estimating the timing of early eukaryotic diversification with multigene molecular clocks".Proceedings of the National Academy of Sciences of the United States of America.108(33): 13624–13629.Bibcode:2011PNAS..10813624P.doi:10.1073/pnas.1110633108.PMC3158185.PMID21810989.
- ^Burki, Fabien; Kaplan, Maia; Tikhonenkov, Denis V.; Zlatogursky, Vasily; Minh, Bui Quang; Radaykina, Liudmila V.; Smirnov, Alexey; Mylnikov, Alexander P.; Keeling, Patrick J. (2016-01-27)."Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista".Proc. R. Soc. B.283(1823): 20152802.doi:10.1098/rspb.2015.2802.ISSN0962-8452.PMC4795036.PMID26817772.
- ^Nishimura, Yuki; Kume, Keitaro; Sonehara, Keito; Tanifuji, Goro; Shiratori, Takashi; Ishida, Ken-Ichiro; Hashimoto, Tetsuo; Inagaki, Yuji; Ohkuma, Moriya (2020)."Mitochondrial Genomes of Hemiarma marina and Leucocryptos marina Revised the Evolution of Cytochrome c Maturation in Cryptista".Frontiers in Ecology and Evolution.8.doi:10.3389/fevo.2020.00140.
- ^"Cryptomonads".Retrieved2009-06-24.