Reverse transcription loop-mediated isothermal amplification(RT-LAMP) is a one step nucleic acid amplification method to multiply specific sequences of RNA. It is used to diagnose infectious disease caused byRNA viruses.[1]
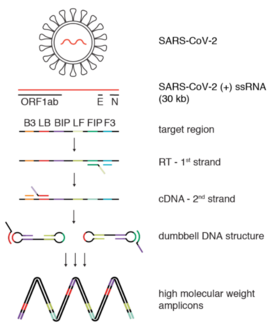
It combinesLAMP[2]DNA-detection withreverse transcription,makingcDNAfrom RNA before running the reaction.[3]RT-LAMP does not requirethermal cycles(unlikePCR) and is performed at a constant temperature between 60 and 65 °C.
RT-LAMP is used in the detection ofRNA viruses(groups II, IV, and V on theBaltimore Virus Classificationsystem), such as theSARS-CoV-2 virus[4]and theEbola virus.[5]
Applications
editRT-LAMP is used to test for the presence of specific RNA-samples of viruses for the specific sequence of the virus, made possible by comparing the sequences against a large external database of references.
Detection of the SARS-CoV2-Virus
editThe RT-LAMP technique is being supported as a cheaper and easier alternative toRT-PCRfor the early diagnostics of people that are infectious forCOVID-19.[6]There areopen accesstest designs (including therecombinant proteins) which makes it legally possible for anyone to produce a test. In contrast to classic rapid tests bylateral flow,RT-LAMP allows the early diagnosis of the disease by testing theviral RNA.[7]
The tests can be done without previous RNA-isolation, detecting the viruses directly from swabs[8]or fromsaliva.[9]
Detection of non-human viruses
editOne example of use case of RT-LAMP was as an experiment to detect a newduckTembusu-like, BYD virus, named after the region,Baiyangdian,where it was first isolated[10][11][1]Another recent application of this method, was in a 2013 experiment to detect an Akabane virus using RT-LAMP. The experiment, done in China, isolated the virus from aborted calf fetuses.[12]
Detection of body fluids
editRT-LAMP is also being used inForensic Serologyto identify body fluids. Researchers have done experiments to show that this method can effectively identify certain body fluids. Knowing there would be limitations, Su et al, come to the conclusion that RT-LAMP was only able to identify blood.[13][14]
Methodology
editReverse transcription
editA specific sequence of the cDNA is detected by 4 LAMPprimers.Two of them areinner primers(FIP and BIP), which serve as base for theBst enzymecopy the template into a new DNA. Theouter primers(F3 and B3) anneal to the template strand and help the reaction to proceed.
As in the case ofRT-PCR,the RT-LAMP procedure starts by making DNA from the sample RNA. This conversion is made by areverse transcriptase,an enzyme derived fromretrovirusescapable of making such a conversion.[15]This DNA derived from RNA is calledcDNA,or complementary DNA. The FIP primer is used by thereverse transcriptaseto build a single-strand of copy DNA. The F3 primer binds to this side of the template strand as well, and displaces the previously made copy.
Amplification
editThis displaced, single-stranded copy is a mixture of target RNA and primers. The primers are designed to have a sequence that binds to the sequence itself, forming a loop.
The BIP primer binds to the other end of this single strand and is used by theBst DNA polymeraseto build a complementary strand, making double-strand DNA. The F3 primer binds to this end and displaces, once again, this newly generatedsingle-stranded DNAmolecule.
This new single strand that has been released will act as the starting point for the LAMP cycling amplification. This single-stranded DNA has adumbbell-like structure as the ends fold and self-bind, forming two loops.
The DNA polymerase and the FIP or BIP primers keep amplifying this strand and the LAMP-reaction product is extended. This cycle can be started from either the forward or backward side of the strand using the appropriate primer. Once this cycle has begun, the strand undergoes self-primed DNA synthesis during the elongation stage of the amplification process. This amplification takes place in less an hour, under isothermal conditions between 60 and 65 °C.
Read out
editThe read out of RT-LAMP tests is frequently colorimetric. Two of the common ways are based on measuring eitherpHormagnesiumions. The amplification reaction causes pH to lower and Mg2+ levels to drop. This can be perceived by indicators, such asPhenol red,for pH, andhydroxynaphthol blue(HNB), for magnesium.[15]Another option is to useSYBR Green I,a DNA intercalating coloring agent.[16]
Advantages and disadvantages
editThis method is specifically advantageous because it can all be done quickly in one step. The sample is mixed with the primers, reverse transcriptase and DNA polymerase and the reaction takes place under a constant temperature. The required temperature can be achieved using a simple hot water bath.
PCR requiresthermocycling;RT-LAMP does not, making it more time efficient and very cost effective.[3]This inexpensive and streamlined method can be more readily used in developing countries that do not have access to high tech laboratories.
A disadvantage of this method is generating the sequence specific primers. For each LAMP assay, primers must be specifically designed to be compatible with the target DNA. This can be difficult which discourages researchers from using the LAMP method in their work.[1]There is however, a free software called Primer Explorer, developed by Fujitsu in Japan, which can aid in the selection of these primers.
See also
editReferences
edit- ^abcMori Y, Notomi T (2009)."Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases".J. Infect. Chemother.15(2): 62–9.doi:10.1007/s10156-009-0669-9.PMC7087713.PMID19396514.
- ^Notomi, Tsugunori; Okayama, Hiroto; Masubuchi, Harumi; Yonekawa, Toshihiro; Watanabe, Keiko; Amino, Nobuyuki; Hase, Tetsu (2000-06-15)."Loop-mediated isothermal amplification of DNA".Nucleic Acids Research.28(12): e63.doi:10.1093/nar/28.12.e63.ISSN0305-1048.PMC102748.PMID10871386.
- ^abFu S, Qu G, Guo S, Ma L, Zhang N, Zhang S, Gao S, Shen Z (2011). "Applications of loop-mediated isothermal DNA amplification".Appl. Biochem. Biotechnol.163(7): 845–50.doi:10.1007/s12010-010-9088-8.PMID20844984.S2CID45682156.
- ^Habibzadeh, Parham; Mofatteh, Mohammad; Silawi, Mohammad; Ghavami, Saeid; Faghihi, Mohammad Ali (2021-02-17)."Molecular diagnostic assays for COVID-19: an overview".Critical Reviews in Clinical Laboratory Sciences.58(6): 385–398.doi:10.1080/10408363.2021.1884640.ISSN1549-781X.PMC7898297.PMID33595397.
- ^Kurosaki, Yohei; Magassouba, N’Faly; Oloniniyi, Olamide K.; Cherif, Mahamoud S.; Sakabe, Saori; Takada, Ayato; Hirayama, Kenji; Yasuda, Jiro (2016-02-22)."Development and Evaluation of Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP) Assay Coupled with a Portable Device for Rapid Diagnosis of Ebola Virus Disease in Guinea".PLOS Neglected Tropical Diseases.10(2): e0004472.doi:10.1371/journal.pntd.0004472.ISSN1935-2735.PMC4764121.PMID26900929.
- ^"LAMP-based Test Could Enable Point-of-Care COVID-19 Testing".Diagnostics from Technology Networks.Retrieved2020-07-31.
- ^Alekseenko, Alisa; Barrett, Donal; Pareja-Sanchez, Yerma; Howard, Rebecca J.; Strandback, Emilia; Ampah-Korsah, Henry; Rovšnik, Urška; Zuniga-Veliz, Silvia; Klenov, Alexander; Malloo, Jayshna; Ye, Shenglong (2021-01-19)."Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples".Scientific Reports.11(1): 1820.Bibcode:2021NatSR..11.1820A.doi:10.1038/s41598-020-80352-8.ISSN2045-2322.PMC7815738.PMID33469065.
- ^Lalli, Matthew A.; Langmade, S Joshua; Chen, Xuhua; Fronick, Catrina C.; Sawyer, Christopher S.; Burcea, Lauren C.; Wilkinson, Michael N.; Fulton, Robert S.; Heinz, Michael; Buchser, William J.; Head, Richard D.; Mitra, Robi D.; Milbrandt, Jeffrey (2020). "Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP".medRxiv10.1101/2020.05.07.20093542.
- ^Nagura-Ikeda, Mayu; Imai, Kazuo; Tabata, Sakiko; Miyoshi, Kazuyasu; Murahara, Nami; Mizuno, Tsukasa; Horiuchi, Midori; Kato, Kento; Imoto, Yoshitaka; Iwata, Maki; Mimura, Satoshi (2020-08-24)."Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription–Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19".Journal of Clinical Microbiology.58(9).doi:10.1128/JCM.01438-20.ISSN0095-1137.PMC7448663.PMID32636214.
- ^Vaidya NK, Wang FB, Zou X, Wahl LM (2012)."Transmission dynamics of the recently-identified BYD virus causing duck egg-drop syndrome".PLOS ONE.7(4): e35161.Bibcode:2012PLoSO...735161V.doi:10.1371/journal.pone.0035161.PMC3329443.PMID22529985.
- ^Jiang T, Liu J, Deng YQ, Su JL, Xu LJ, Liu ZH, Li XF, Yu XD, Zhu SY, Gao GF, Qin ED, Qin CF (December 2012). "Development of RT-LAMP and real-time RT-PCR assays for the rapid detection of the new duck Tembusu-like BYD virus".Arch. Virol.157(12): 2273–80.doi:10.1007/s00705-012-1431-7.PMID22865206.S2CID15573433.
- ^Qiao J, Wang J, Meng Q, Wang G, Liu Y, He Z, Yang H, Zhang Z, Cai X, Chen C (2013)."Rapid detection of Akabane virus by a novel reverse transcription loop-mediated isothermal amplification assay (RT-LAMP)".Virol. J.10:288.doi:10.1186/1743-422X-10-288.PMC3848447.PMID24034624.
- ^Su, Chih-Wen; Li, Chiao-Yun; Lee, James Chun-I; Ji, Dar-Der; Li, Shu-Ying; Daniel, Barbara; Syndercombe-Court, Denise; Linacre, Adrian; Hsieh, Hsing-Mei (2015-06-01)."A novel application of real-time RT-LAMP for body fluid identification: using HBB detection as the model".Forensic Science, Medicine, and Pathology.11(2): 208–215.doi:10.1007/s12024-015-9668-6.ISSN1556-2891.PMID25877518.S2CID19288327.
- ^Satoh, Tetsuya; Kouroki, Seiya; Ogawa, Keita; Tanaka, Yorika; Matsumura, Kazutoshi; Iwase, Susumu (2018-07-01)."Development of mRNA-based body fluid identification using reverse transcription loop-mediated isothermal amplification".Analytical and Bioanalytical Chemistry.410(18): 4371–4378.doi:10.1007/s00216-018-1088-5.ISSN1618-2650.PMID29696299.S2CID13823864.
- ^abKellner, Max J.; Ross, James J.; Schnabl, Jakob; Dekens, Marcus P. S.; Heinen, Robert; Grishkovskaya, Irina; Bauer, Benedikt; Stadlmann, Johannes; Menéndez-Arias, Luis; Fritsche-Polanz, Robert; Traugott, Marianna (2020-07-23)."A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing".bioRxiv:2020.06.23.166397.doi:10.1101/2020.06.23.166397.hdl:10261/216969.S2CID220835822.
- ^Bokelmann, Lukas; Nickel, Olaf; Maricic, Tomislav; Paabo, Svante; Meyer, Matthias; Borte, Stephan; Riesenberg, Stephan (2020-08-06). "Rapid, reliable, and cheap point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP (Cap-iLAMP)".medRxiv10.1101/2020.08.04.20168617.