Ribosomes(/ˈraɪbəzoʊm,-soʊm/) aremacromolecular machines,found within allcells,that performbiological protein synthesis(messenger RNAtranslation). Ribosomes linkamino acidstogether in the order specified by thecodonsof messenger RNA molecules to formpolypeptidechains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or moreribosomal RNAmolecules and manyribosomal proteins(r-proteins).[1][2][3]The ribosomes and associated molecules are also known as thetranslational apparatus.
| Cell biology | |
|---|---|
| Animal cell diagram | |
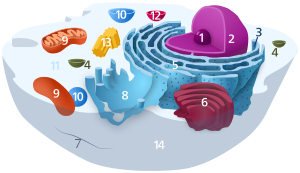 Components of a typical animal cell:
|
Overview
editThe sequence ofDNAthat encodes the sequence of theamino acidsin a protein is transcribed into amessenger RNA(mRNA) chain. Ribosomes bind to the messenger RNA molecules and use the RNA's sequence ofnucleotidesto determine the sequence of amino acids needed to generate a protein. Amino acids are selected and carried to the ribosome bytransfer RNA(tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via ananti-codonstem loop. For each coding triplet (codon) in the messenger RNA, there is a unique transfer RNA that must have the exact anti-codon match, and carries the correct amino acid for incorporating into a growingpolypeptidechain. Once the protein is produced, it can thenfoldto produce a functional three-dimensional structure.
A ribosome is made fromcomplexesof RNAs and proteins and is therefore aribonucleoprotein complex.In prokaryotes each ribosome is composed of small (30S) and large (50S) components, called subunits, which are bound to each other:
- (30S) has mainly a decoding function and is also bound to the mRNA
- (50S) has mainly a catalytic function and is also bound to the aminoacylated tRNAs.
The synthesis of proteins from their building blocks takes place in four phases: initiation, elongation, termination, and recycling. The start codon in all mRNA molecules has the sequence AUG. The stop codon is one of UAA, UAG, or UGA; since there are no tRNA molecules that recognize these codons, the ribosome recognizes that translation is complete.[4]When a ribosome finishes reading an mRNA molecule, the two subunits separate and are usually broken up but can be reused. Ribosomes are a kind ofenzyme,calledribozymesbecause thecatalyticpeptidyl transferaseactivity that links amino acids together is performed by the ribosomal RNA.[5]
Ineukaryotic cells,ribosomes are often associated with the intracellular membranes that make up therough endoplasmic reticulum.
Ribosomes frombacteria,archaea,andeukaryotes(in thethree-domain system) resemble each other to a remarkable degree, evidence of a common origin. They differ in their size, sequence, structure, and the ratio of protein to RNA. The differences in structure allow someantibioticsto kill bacteria by inhibiting their ribosomes while leaving human ribosomes unaffected. In all species, more than one ribosome may move along a single mRNA chain at one time (as apolysome), each "reading" a specific sequence and producing a corresponding protein molecule.
Themitochondrial ribosomesof eukaryotic cells are distinct from their other ribosomes. They functionally resemble those in bacteria, reflecting the evolutionary origin of mitochondria asendosymbioticbacteria.[6][7]
Discovery
editRibosomes were first observed in the mid-1950s byRomanian-Americancell biologistGeorge Emil Palade,using anelectron microscope,as dense particles or granules.[8]They were initially calledPalade granulesdue to their granular structure. The term "ribosome" was proposed in 1958 by Howard M. Dintzis:[9]
During the course of the symposium a semantic difficulty became apparent. To some of the participants, "microsomes" mean the ribonucleoprotein particles of the microsome fraction contaminated by other protein and lipid material; to others, the microsomes consist of protein and lipid contaminated by particles. The phrase "microsomal particles" does not seem adequate, and "ribonucleoprotein particles of the microsome fraction" is much too awkward. During the meeting, the word "ribosome" was suggested, which has a very satisfactory name and a pleasant sound. The present confusion would be eliminated if "ribosome" were adopted to designate ribonucleoprotein particles in sizes ranging from 35 to 100S.
— Albert Claude, Microsomal Particles and Protein Synthesis[10]
Albert Claude,Christian de Duve,andGeorge Emil Paladewere jointly awarded theNobel Prize in Physiology or Medicine,in 1974, for the discovery of the ribosome.[11]TheNobel PrizeinChemistry2009 was awarded toVenkatraman Ramakrishnan,Thomas A. SteitzandAda E. Yonathfor determining the detailed structure and mechanism of the ribosome.[12]
Structure
editThe ribosome is a complex cellular machine. It is largely made up of specialized RNA known asribosomal RNA(rRNA) as well as dozens of distinct proteins (the exact number varies slightly between species). The ribosomal proteins and rRNAs are arranged into two distinct ribosomal pieces of different sizes, known generally as the large and small subunits of the ribosome. Ribosomes consist of two subunits that fit together and work as one to translate the mRNA into a polypeptide chain during protein synthesis. Because they are formed from two subunits of non-equal size, they are slightly longer on the axis than in diameter.
Prokaryotic ribosomes
editProkaryotic ribosomes are around 20nm(200Å) in diameter and are composed of 65% rRNA and 35%ribosomal proteins.[13]Eukaryotic ribosomes are between 25 and 30nm(250–300 Å) in diameter with an rRNA-to-protein ratio that is close to 1.[14]Crystallographicwork[15]has shown that there are no ribosomal proteins close to the reaction site for polypeptide synthesis. This suggests that the protein components of ribosomes do not directly participate in peptide bond formation catalysis, but rather that these proteins act as a scaffold that may enhance the ability of rRNA to synthesize protein (see:Ribozyme).
The ribosomal subunits ofprokaryotesandeukaryotesare quite similar.[17]
The unit of measurement used to describe the ribosomal subunits and the rRNA fragments is theSvedbergunit, a measure of the rate ofsedimentationincentrifugationrather than size. This accounts for why fragment names do not add up: for example, bacterial 70S ribosomes are made of 50S and 30S subunits.
Prokaryotes have 70Sribosomes, each consisting of a small (30S) and a large (50S) subunit.E. coli,for example, has a16SRNA subunit (consisting of 1540 nucleotides) that is bound to 21 proteins. The large subunit is composed of a5SRNA subunit (120 nucleotides), a 23S RNA subunit (2900 nucleotides) and 31proteins.[17]
Ribosome ofE. coli(a bacterium)[18]: 962 ribosome subunit rRNAs r-proteins 70S 50S 23S (2904nt) 31 5S (120 nt) 30S 16S (1542 nt) 21
Affinity label for the tRNA binding sites on theE. coliribosome allowed the identification of A and P site proteins most likely associated with the peptidyltransferase activity;[5]labelled proteins are L27, L14, L15, L16, L2; at least L27 is located at the donor site, as shown by E. Collatz and A.P. Czernilofsky.[19][20]Additional research has demonstrated that the S1 and S21 proteins, in association with the 3′-end of 16S ribosomal RNA, are involved in the initiation of translation.[21]
Archaeal ribosomes
editArchaeal ribosomes share the same general dimensions of bacteria ones, being a 70S ribosome made up from a 50S large subunit, a 30S small subunit, and containing three rRNA chains. However, on the sequence level, they are much closer to eukaryotic ones than to bacterial ones. Every extra ribosomal protein archaea have compared to bacteria has a eukaryotic counterpart, while no such relation applies between archaea and bacteria.[22][23][24]
Eukaryotic ribosomes
editEukaryotes have 80S ribosomes located in their cytosol, each consisting of asmall (40S)andlarge (60S) subunit.Their 40S subunit has an18S RNA(1900 nucleotides) and 33 proteins.[25][26]The large subunit is composed of a5S RNA(120 nucleotides),28S RNA(4700 nucleotides), a5.8S RNA(160 nucleotides) subunits and 49 proteins.[17][25][27]
eukaryotic cytosolic ribosomes (R. norvegicus)[18]: 65 ribosome subunit rRNAs r-proteins 80S 60S 28S (4718 nt) 49 5.8S (160 nt) 5S (120 nt) 40S 18S (1874 nt) 33
During 1977, Czernilofsky published research that usedaffinity labelingto identify tRNA-binding sites on rat liver ribosomes. Several proteins, including L32/33, L36, L21, L23, L28/29 and L13 were implicated as being at or near thepeptidyl transferasecenter.[28]
Plastoribosomes and mitoribosomes
editIn eukaryotes, ribosomes are present inmitochondria(sometimes calledmitoribosomes) and inplastidssuch aschloroplasts(also called plastoribosomes). They also consist of large and small subunits bound together withproteinsinto one 70S particle.[17]These ribosomes are similar to those of bacteria and these organelles are thought to have originated assymbiotic bacteria.[17]Of the two, chloroplastic ribosomes are closer to bacterial ones than mitochondrial ones are. Many pieces of ribosomal RNA in the mitochondria are shortened, and in the case of5S rRNA,replaced by other structures in animals and fungi.[29]In particular,Leishmaniatarentolaehas a minimalized set of mitochondrial rRNA.[30]In contrast, plant mitoribosomes have both extended rRNA and additional proteins as compared to bacteria, in particular, many pentatricopetide repeat proteins.[31]
Thecryptomonadandchlorarachniophytealgae may contain anucleomorphthat resembles a vestigial eukaryotic nucleus.[32]Eukaryotic 80S ribosomes may be present in the compartment containing the nucleomorph.[33]
Making use of the differences
editThe differences between the bacterial and eukaryotic ribosomes are exploited bypharmaceutical chemiststo createantibioticsthat can destroy a bacterial infection without harming the cells of the infected person. Due to the differences in their structures, the bacterial 70S ribosomes are vulnerable to these antibiotics while the eukaryotic 80S ribosomes are not.[34]Even thoughmitochondriapossess ribosomes similar to the bacterial ones, mitochondria are not affected by these antibiotics because they are surrounded by a double membrane that does not easily admit these antibiotics into theorganelle.[35]A noteworthy counterexample is the antineoplastic antibioticchloramphenicol,which inhibits bacterial 50S and eukaryotic mitochondrial 50S ribosomes.[36]Ribosomes in chloroplasts, however, are different: Antibiotic resistance in chloroplast ribosomal proteins is a trait that has to be introduced as a marker, with genetic engineering.[37]
Common properties
editThe various ribosomes share a core structure, which is quite similar despite the large differences in size. Much of the RNA is highly organized into varioustertiary structural motifs,for examplepseudoknotsthat exhibitcoaxial stacking.The extraRNAin the larger ribosomes is in several long continuous insertions,[38]such that they form loops out of the core structure without disrupting or changing it.[17]All of the catalytic activity of the ribosome is carried out by theRNA;the proteins reside on the surface and seem to stabilize the structure.[17]
High-resolution structure
editThe general molecular structure of the ribosome has been known since the early 1970s. In the early 2000s, the structure has been achieved at high resolutions, of the order of a fewångströms.
The first papers giving the structure of the ribosome at atomic resolution were published almost simultaneously in late 2000. The 50S (large prokaryotic) subunit was determined from thearchaeonHaloarcula marismortui[39]and thebacteriumDeinococcus radiodurans,and the structure of the 30S subunit was determined from the bacteriumThermus thermophilus.[16][40]These structural studies were awarded the Nobel Prize in Chemistry in 2009. In May 2001 these coordinates were used to reconstruct the entireT. thermophilus70S particle at 5.5Åresolution.[41]
Two papers were published in November 2005 with structures of theEscherichia coli70S ribosome. The structures of a vacant ribosome were determined at 3.5Åresolution usingX-ray crystallography.[42]Then, two weeks later, a structure based oncryo-electron microscopywas published,[43]which depicts the ribosome at 11–15Åresolution in the act of passing a newly synthesized protein strand into the protein-conducting channel.
The first atomic structures of the ribosome complexed withtRNAandmRNAmolecules were solved by using X-ray crystallography by two groups independently, at 2.8Å[44]and at 3.7Å.[45]These structures allow one to see the details of interactions of theThermus thermophilusribosome withmRNAand withtRNAsbound at classical ribosomal sites. Interactions of the ribosome with long mRNAs containingShine-Dalgarno sequenceswere visualized soon after that at 4.5–5.5Åresolution.[46]
In 2011, the first complete atomic structure of the eukaryotic 80S ribosome from the yeastSaccharomyces cerevisiaewas obtained by crystallography.[25]The model reveals the architecture of eukaryote-specific elements and their interaction with the universally conserved core. At the same time, the complete model of a eukaryotic 40S ribosomal structure inTetrahymena thermophilawas published and described the structure of the40S subunit,as well as much about the 40S subunit's interaction witheIF1duringtranslation initiation.[26]Similarly, the eukaryotic60S subunitstructure was also determined fromTetrahymena thermophilain complex witheIF6.[27]
Function
editRibosomes are minute particles consisting of RNA and associated proteins that function to synthesize proteins. Proteins are needed for many cellular functions, such as repairing damage or directing chemical processes. Ribosomes can be found floating within the cytoplasm or attached to theendoplasmic reticulum.Their main function is to convert genetic code into an amino acid sequence and to build protein polymers from amino acid monomers.
Ribosomes act as catalysts in two extremely important biological processes called peptidyl transfer and peptidyl hydrolysis.[5][47]The "PT center is responsible for producing protein bonds during protein elongation".[47]
In summary, ribosomes have two main functions: Decoding the message, and the formation of peptide bonds. These two functions reside in the ribosomal subunits. Each subunit is made of one or more rRNAs and many r-proteins. The small subunit (30S in bacteria and archaea, 40S in eukaryotes) has the decoding function, whereas the large subunit (50S in bacteria and archaea, 60S in eukaryotes) catalyzes the formation of peptide bonds, referred to as the peptidyl-transferase activity. The bacterial (and archaeal) small subunit contains the 16S rRNA and 21 r-proteins (Escherichia coli), whereas the eukaryotic small subunit contains the 18S rRNA and 32 r-proteins (Saccharomyces cerevisiae, although the numbers vary between species). The bacterial large subunit contains the 5S and 23S rRNAs and 34 r-proteins (E. coli), with the eukaryotic large subunit containing the 5S, 5.8S, and 25S/28S rRNAs and 46 r-proteins (S. cerevisiae;again, the exact numbers vary between species).[48]
Translation
editRibosomes are the workplaces ofprotein biosynthesis,the process of translatingmRNAintoprotein.The mRNA comprises a series ofcodonswhich are decoded by the ribosome to make the protein. Using the mRNA as a template, the ribosome traverses each codon (3nucleotides) of the mRNA, pairing it with the appropriate amino acid provided by anaminoacyl-tRNA.Aminoacyl-tRNA contains a complementaryanticodonon one end and the appropriate amino acid on the other. For fast and accurate recognition of the appropriate tRNA, the ribosome utilizes large conformational changes (conformational proofreading).[49]The small ribosomal subunit, typically bound to an aminoacyl-tRNA containing the first amino acidmethionine,binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The ribosome contains three RNA binding sites, designated A, P, and E. TheA-sitebinds an aminoacyl-tRNA or termination release factors;[50][51]theP-sitebinds a peptidyl-tRNA (a tRNA bound to the poly-peptide chain); and theE-site(exit) binds a free tRNA. Protein synthesis begins at astart codonAUG near the 5' end of the mRNA. mRNA binds to the P site of the ribosome first. The ribosome recognizes the start codon by using theShine-Dalgarno sequenceof the mRNA in prokaryotes andKozak boxin eukaryotes.
Although catalysis of thepeptide bondinvolves the C2hydroxylof RNA's P-siteadenosinein a proton shuttle mechanism, other steps in protein synthesis (such as translocation) are caused by changes in protein conformations. Since theircatalytic coreis made of RNA, ribosomes are classified as "ribozymes,"[52]and it is thought that they might be remnants of theRNA world.[53]
In Figure 5, both ribosomal subunits (smallandlarge) assemble at the start codon (towards the 5' end of themRNA). The ribosome usestRNAthat matches the current codon (triplet) on the mRNA to append anamino acidto the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves towards the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what is called apolyribosomeorpolysome.
Cotranslational folding
editThe ribosome is known to actively participate in theprotein folding.[54][55]The structures obtained in this way are usually identical to the ones obtained during protein chemical refolding; however, the pathways leading to the final product may be different.[56][57]In some cases, the ribosome is crucial in obtaining the functional protein form. For example, one of the possible mechanisms of folding of the deeplyknotted proteinsrelies on the ribosome pushing the chain through the attached loop.[58]
Addition of translation-independent amino acids
editPresence of a ribosome quality control protein Rqc2 is associated with mRNA-independent protein elongation.[59][60]This elongation is a result of ribosomal addition (via tRNAs brought by Rqc2) ofCATtails:ribosomes extend theC-terminusof a stalled protein with random, translation-independent sequences ofalaninesandthreonines.[61][62]
Ribosome locations
editRibosomes are classified as being either "free" or "membrane-bound".
Free and membrane-bound ribosomes differ only in their spatial distribution; they are identical in structure. Whether the ribosome exists in a free or membrane-bound state depends on the presence of anER-targeting signal sequenceon the protein being synthesized, so an individual ribosome might be membrane-bound when it is making one protein, but free in the cytosol when it makes another protein.
Ribosomes are sometimes referred to asorganelles,but the use of the termorganelleis often restricted to describing sub-cellular components that include a phospholipid membrane, which ribosomes, being entirely particulate, do not. For this reason, ribosomes may sometimes be described as "non-membranous organelles".
Free ribosomes
editFree ribosomes can move about anywhere in thecytosol,but are excluded from thecell nucleusand other organelles. Proteins that are formed from free ribosomes are released into the cytosol and used within the cell. Since the cytosol contains high concentrations ofglutathioneand is, therefore, areducing environment,proteins containingdisulfide bonds,which are formed from oxidized cysteine residues, cannot be produced within it.
Membrane-bound ribosomes
editWhen a ribosome begins to synthesize proteins that are needed in some organelles, the ribosome making this protein can become "membrane-bound". In eukaryotic cells this happens in a region of the endoplasmic reticulum (ER) called the "rough ER". The newly produced polypeptide chains are inserted directly into the ER by the ribosome undertakingvectorial synthesisand are then transported to their destinations, through thesecretory pathway.Bound ribosomes usually produce proteins that are used within the plasma membrane or are expelled from the cell viaexocytosis.[63]
Biogenesis
editIn bacterial cells, ribosomes are synthesized in the cytoplasm through thetranscriptionof multiple ribosome geneoperons.In eukaryotes, the process takes place both in the cell cytoplasm and in thenucleolus,which is a region within thecell nucleus.The assembly process involves the coordinated function of over 200 proteins in the synthesis and processing of the four rRNAs, as well as assembly of those rRNAs with the ribosomal proteins.[64]
Origin
editThe ribosome may have first originated as a protoribosome,[65]possibly containing a peptidyl transferase centre (PTC), in anRNA world,appearing as a self-replicating complex that only later evolved the ability to synthesize proteins whenamino acidsbegan to appear.[66]Studies suggest that ancient ribosomes constructed solely ofrRNAcould have developed the ability to synthesizepeptide bonds.[67][68][69][70][71]In addition, evidence strongly points to ancient ribosomes as self-replicating complexes, where the rRNA in the ribosomes had informational, structural, and catalytic purposes because it could have coded fortRNAsand proteins needed for ribosomal self-replication.[72]Hypothetical cellular organisms with self-replicating RNA but without DNA are calledribocytes(or ribocells).[73][74]
As amino acids gradually appeared in the RNA world under prebiotic conditions,[75][76]their interactions with catalytic RNA would increase both the range and efficiency of function of catalytic RNA molecules.[66]Thus, the driving force for the evolution of the ribosome from an ancientself-replicating machineinto its current form as a translational machine may have been the selective pressure to incorporate proteins into the ribosome's self-replicating mechanisms, so as to increase its capacity for self-replication.[72][77][78]
Heterogeneous ribosomes
editRibosomes are compositionally heterogeneous between species and even within the same cell, as evidenced by the existence of cytoplasmic and mitochondria ribosomes within the same eukaryotic cells. Certain researchers have suggested that heterogeneity in the composition of ribosomal proteins in mammals is important for gene regulation,i.e.,the specialized ribosome hypothesis.[79][80]However, this hypothesis is controversial and the topic of ongoing research.[81][82]
Heterogeneity in ribosome composition was first proposed to be involved in translational control of protein synthesis by Vince Mauro andGerald Edelman.[83]They proposed the ribosome filter hypothesis to explain the regulatory functions of ribosomes. Evidence has suggested that specialized ribosomes specific to different cell populations may affect how genes are translated.[84]Some ribosomal proteins exchange from the assembled complex withcytosoliccopies[85]suggesting that the structure of thein vivoribosome can be modified without synthesizing an entire new ribosome.
Certain ribosomal proteins are absolutely critical for cellular life while others are not. Inbudding yeast,14/78 ribosomal proteins are non-essential for growth, while in humans this depends on the cell of study.[86]Other forms of heterogeneity include post-translational modifications to ribosomal proteins such as acetylation, methylation, and phosphorylation.[87]Arabidopsis,[88][89][90][91]Viralinternal ribosome entry sites (IRESs)may mediate translations by compositionally distinct ribosomes. For example, 40S ribosomal units withouteS25in yeast and mammalian cells are unable to recruit theCrPV IGR IRES.[92]
Heterogeneity of ribosomal RNA modifications plays a significant role in structural maintenance and/or function and most mRNA modifications are found in highly conserved regions.[93][94]The most common rRNA modifications arepseudouridylationand2'-O-methylationof ribose.[95]
See also
edit- Aminoglycosides
- Biological machines
- Posttranslational modification
- Protein dynamics
- Ribosome-associated vesicle
- RNA tertiary structure
- Translation (genetics)
- Wobble base pair
- Ada Yonath—Israeli crystallographer known for her pioneering work on the structure of the ribosome, for which she won theNobel Prize.
References
edit- ^Konikkat S (February 2016).Dynamic Remodeling Events Drive the Removal of the ITS2 Spacer Sequence During Assembly of 60S Ribosomal Subunits in S. cerevisiae(Ph.D. thesis). Carnegie Mellon University. Archived fromthe originalon 3 August 2017.
- ^Weiler EW, Nover L (2008).Allgemeine und Molekulare Botanik(in German). Stuttgart: Georg Thieme Verlag. p. 532.ISBN9783131527912.
- ^de la Cruz J, Karbstein K, Woolford JL (2015)."Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo".Annual Review of Biochemistry.84:93–129.doi:10.1146/annurev-biochem-060614-033917.PMC4772166.PMID25706898.
- ^"Scitable by nature translation / RNA translation".
- ^abcTirumalai MR, Rivas M, Tran Q, Fox GE (November 2021)."The Peptidyl Transferase Center: a Window to the Past".Microbiol Mol Biol Rev.85(4): e0010421.doi:10.1128/MMBR.00104-21.PMC8579967.PMID34756086.
- ^Benne R, Sloof P (1987). "Evolution of the mitochondrial protein synthetic machinery".Bio Systems.21(1): 51–68.Bibcode:1987BiSys..21...51B.doi:10.1016/0303-2647(87)90006-2.PMID2446672.
- ^"Ribosomes".Archived fromthe originalon 2009-03-20.Retrieved2011-04-28.
- ^Palade GE (January 1955)."A small particulate component of the cytoplasm".The Journal of Biophysical and Biochemical Cytology.1(1): 59–68.doi:10.1083/jcb.1.1.59.PMC2223592.PMID14381428.
- ^Rheinberger, Hans-Jörg (September 2022)."A Brief History of Protein Biosynthesis and Ribosome Research".Lindau Nobel Laureate Meetings.Retrieved2023-08-16.
- ^Roberts RB, ed. (1958). "Introduction".Microsomal Particles and Protein Synthesis.New York: Pergamon Press, Inc.
- ^"The Nobel Prize in Physiology or Medicine 1974".Nobelprize.org.The Nobel Foundation.Archivedfrom the original on 26 January 2013.Retrieved10 December2012.
- ^"2009 Nobel Prize in Chemistry".The Nobel Foundation.Archivedfrom the original on 28 April 2012.Retrieved10 December2012.
- ^Kurland CG(1960). "Molecular characterization of ribonucleic acid from Escherichia coli ribosomes".Journal of Molecular Biology.2(2): 83–91.doi:10.1016/s0022-2836(60)80029-0.
- ^Wilson DN, Doudna Cate JH (May 2012)."The structure and function of the eukaryotic ribosome".Cold Spring Harbor Perspectives in Biology.4(5): a011536.doi:10.1101/cshperspect.a011536.PMC3331703.PMID22550233.
- ^Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (August 2000)."The structural basis of ribosome activity in peptide bond synthesis"(PDF).Science.289(5481): 920–30.Bibcode:2000Sci...289..920N.doi:10.1126/science.289.5481.920.PMID10937990.S2CID8370119.Archived fromthe original(PDF)on 2020-11-30.
- ^abWimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (September 2000). "Structure of the 30S ribosomal subunit".Nature.407(6802): 327–39.Bibcode:2000Natur.407..327W.doi:10.1038/35030006.PMID11014182.S2CID4419944.
- ^abcdefgAlberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002)."Membrane-bound Ribosomes Define the Rough ER".Molecular Biology of the Cell(4th ed.). New York: Garland Science. p. 342.ISBN978-0-8153-4072-0.
- ^abGarrett R, Grisham CM (2009).Biochemistry(4th ed.). Cengage Learning Services.ISBN978-0-495-11464-2.
- ^Collatz E, Küchler E, Stöffler G, Czernilofsky AP (April 1976)."The site of reaction on ribosomal protein L27 with an affinity label derivative of tRNA Met f".FEBS Letters.63(2): 283–6.doi:10.1016/0014-5793(76)80112-3.PMID770196.
- ^Czernilofsky AP, Collatz EE, Stöffler G, Kuechler E (January 1974)."Proteins at the tRNA binding sites of Escherichia coli ribosomes".Proceedings of the National Academy of Sciences of the United States of America.71(1): 230–4.Bibcode:1974PNAS...71..230C.doi:10.1073/pnas.71.1.230.PMC387971.PMID4589893.
- ^Czernilofsky AP, Kurland CG, Stöffler G (October 1975)."30S ribosomal proteins associated with the 3'-terminus of 16S RNA".FEBS Letters.58(1): 281–4.doi:10.1016/0014-5793(75)80279-1.PMID1225593.
- ^Cullen KE (2009). "Archaeal Ribosomes".Encyclopedia of Life Science.New York: Facts On File. pp. 1–5.doi:10.1002/9780470015902.a0000293.pub3.ISBN9780470015902.S2CID243730576.
- ^Tirumalai MR, Anane-Bediakoh D, Rajesh R, Fox GE (November 2021)."Net Charges of the Ribosomal Proteins of theS10andspcClusters of Halophiles Are Inversely Related to the Degree of Halotolerance ".Microbiol. Spectr.9(3): e0178221.doi:10.1128/spectrum.01782-21.PMC8672879.PMID34908470.
- ^Wang J, Dasgupta I, Fox GE (28 April 2009)."Many nonuniversal archaeal ribosomal proteins are found in conserved gene clusters".Archaea.2(4): 241–51.doi:10.1155/2009/971494.PMC2686390.PMID19478915.
- ^abcBen-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (December 2011)."The structure of the eukaryotic ribosome at 3.0 Å resolution".Science.334(6062): 1524–9.Bibcode:2011Sci...334.1524B.doi:10.1126/science.1212642.PMID22096102.S2CID9099683.
- ^abRabl J, Leibundgut M, Ataide SF, Haag A, Ban N (February 2011)."Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1"(PDF).Science.331(6018): 730–6.Bibcode:2011Sci...331..730R.doi:10.1126/science.1198308.hdl:20.500.11850/153130.PMID21205638.S2CID24771575.
- ^abKlinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (November 2011). "Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6".Science.334(6058): 941–8.Bibcode:2011Sci...334..941K.doi:10.1126/science.1211204.PMID22052974.S2CID206536444.
- ^Fabijanski S, Pellegrini M (1977). "Identification of proteins at the peptidyl-tRNA binding site of rat liver ribosomes".Molecular & General Genetics.184(3): 551–6.doi:10.1007/BF00431588.PMID6950200.S2CID9751945.
- ^Agrawal RK, Sharma MR (December 2012)."Structural aspects of mitochondrial translational apparatus".Current Opinion in Structural Biology.22(6): 797–803.doi:10.1016/j.sbi.2012.08.003.PMC3513651.PMID22959417.
- ^Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK (June 2009)."Structure of a mitochondrial ribosome with minimal RNA".Proceedings of the National Academy of Sciences of the United States of America.106(24): 9637–42.Bibcode:2009PNAS..106.9637S.doi:10.1073/pnas.0901631106.PMC2700991.PMID19497863.
- ^Waltz F, Nguyen TT, Arrivé M, Bochler A, Chicher J, Hammann P, Kuhn L, Quadrado M, Mireau H, Hashem Y, Giegé P (January 2019). "Small is big in Arabidopsis mitochondrial ribosome".Nature Plants.5(1): 106–117.doi:10.1038/s41477-018-0339-y.PMID30626926.S2CID58004990.
- ^Archibald JM, Lane CE (2009)."Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction".The Journal of Heredity.100(5): 582–90.doi:10.1093/jhered/esp055.PMID19617523.
- ^"Specialized Internal Structures of Prokaryotes | Boundless Microbiology".courses.lumenlearning.Retrieved2021-09-24.
- ^Recht MI, Douthwaite S, Puglisi JD (June 1999)."Basis for prokaryotic specificity of action of aminoglycoside antibiotics".The EMBO Journal.18(11): 3133–8.doi:10.1093/emboj/18.11.3133.PMC1171394.PMID10357824.
- ^O'Brien TW (May 1971)."The general occurrence of 55 S ribosomes in mammalian liver mitochondria".The Journal of Biological Chemistry.246(10): 3409–17.doi:10.1016/S0021-9258(18)62239-2.PMID4930061.
- ^"Chloramphenicol-lnduced Bone Marrow Suppression".JAMA.213(7): 1183–1184. 1970-08-17.doi:10.1001/jama.1970.03170330063011.ISSN0098-7484.PMID5468266.
- ^Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH (December 1990)."Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events".Genetics.126(4): 875–88.doi:10.1093/genetics/126.4.875.PMC1204285.PMID1981764.
- ^Penev PI, Fakhretaha-Aval S, Patel VJ, Cannone JJ, Gutell RR, Petrov AS, Williams LD, Glass JB (August 2020)."Supersized ribosomal RNA expansion segments in Asgard archaea".Genome Biology and Evolution.12(10): 1694–1710.doi:10.1093/gbe/evaa170.PMC7594248.PMID32785681.
- ^abBan N, Nissen P, Hansen J, Moore PB, Steitz TA (August 2000). "The complete atomic structure of the large ribosomal subunit at 2.4 A resolution".Science.289(5481): 905–20.Bibcode:2000Sci...289..905B.CiteSeerX10.1.1.58.2271.doi:10.1126/science.289.5481.905.PMID10937989.
- ^Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A (September 2000)."Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution".Cell.102(5): 615–23.doi:10.1016/S0092-8674(00)00084-2.PMID11007480.S2CID1024446.
- ^Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (May 2001)."Crystal structure of the ribosome at 5.5 A resolution".Science.292(5518): 883–96.Bibcode:2001Sci...292..883Y.doi:10.1126/science.1060089.PMID11283358.S2CID39505192.
- ^Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (November 2005). "Structures of the bacterial ribosome at 3.5 A resolution".Science.310(5749): 827–34.Bibcode:2005Sci...310..827S.doi:10.1126/science.1117230.PMID16272117.S2CID37382005.
- ^Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J (November 2005)."Structure of the E. coli protein-conducting channel bound to a translating ribosome".Nature.438(7066): 318–24.Bibcode:2005Natur.438..318M.doi:10.1038/nature04133.PMC1351281.PMID16292303.
- ^Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (September 2006). "Structure of the 70S ribosome complexed with mRNA and tRNA".Science.313(5795): 1935–42.Bibcode:2006Sci...313.1935S.doi:10.1126/science.1131127.PMID16959973.S2CID9737925.
- ^Korostelev A, Trakhanov S, Laurberg M, Noller HF (September 2006)."Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements".Cell.126(6): 1065–77.doi:10.1016/j.cell.2006.08.032.PMID16962654.S2CID13452915.
- ^Yusupova G, Jenner L, Rees B, Moras D, Yusupov M (November 2006). "Structural basis for messenger RNA movement on the ribosome".Nature.444(7117): 391–4.Bibcode:2006Natur.444..391Y.doi:10.1038/nature05281.PMID17051149.S2CID4419198.
- ^ab"Specialized Internal Structures of Prokaryotes".courses.lumenlearning.Boundless Microbiology.Retrieved2018-09-27.
- ^Lafontaine, D.; Tollervey, D. (2001). "The function and synthesis of ribosomes".Nat Rev Mol Cell Biol.2(7): 514–520.doi:10.1038/35080045.hdl:1842/729.PMID11433365.S2CID2637106.
- ^Savir Y, Tlusty T (April 2013)."The ribosome as an optimal decoder: A lesson in molecular recognition".Cell.153(2): 471–479.Bibcode:2013APS..MARY46006T.doi:10.1016/j.cell.2013.03.032.PMID23582332.
- ^Korkmaz G, Sanyal S (September 2017)."Escherichia coli".The Journal of Biological Chemistry.292(36): 15134–15142.doi:10.1074/jbc.M117.785238.PMC5592688.PMID28743745.
- ^Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI (January 2004)."Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking andMg2+-dependentinteractions ".RNA.10(1): 90–101.doi:10.1261/rna.5142404.PMC1370521.PMID14681588.
- ^Rodnina MV, Beringer M, Wintermeyer W (January 2007). "How ribosomes make peptide bonds".Trends in Biochemical Sciences.32(1): 20–26.doi:10.1016/j.tibs.2006.11.007.PMID17157507.
- ^Cech, T.R. (August 2000). "Structural biology. The ribosome is a ribozyme".Science.289(5481): 878–879.doi:10.1126/science.289.5481.878.PMID10960319.S2CID24172338.
- ^Banerjee D, Sanyal S (October 2014)."Protein folding activity of the ribosome (PFAR) – a target for antiprion compounds".Viruses.6(10): 3907–3924.doi:10.3390/v6103907.PMC4213570.PMID25341659.
- ^Fedorov AN, Baldwin TO (December 1997)."Cotranslational protein folding".The Journal of Biological Chemistry.272(52): 32715–32718.doi:10.1074/jbc.272.52.32715.PMID9407040.
- ^Baldwin RL (June 1975). "Intermediates in protein folding reactions and the mechanism of protein folding".Annual Review of Biochemistry.44(1): 453–475.doi:10.1146/annurev.bi.44.070175.002321.PMID1094916.
- ^Das D, Das A, Samanta D, Ghosh J, Dasgupta S, Bhattacharya A, Basu A, Sanyal S, Das Gupta C (August 2008)."Role of the ribosome in protein folding"(PDF).Biotechnology Journal.3(8): 999–1009.doi:10.1002/biot.200800098.PMID18702035.
- ^Dabrowski-Tumanski P, Piejko M, Niewieczerzal S, Stasiak A, Sulkowska JI (December 2018). "Protein knotting by active threading of nascent polypeptide chain exiting from the ribosome exit channel".The Journal of Physical Chemistry B.122(49): 11616–11625.doi:10.1021/acs.jpcb.8b07634.PMID30198720.S2CID52176392.
- ^Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS (November 2012)."A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress".Cell.151(5): 1042–1054.doi:10.1016/j.cell.2012.10.044.PMC3534965.PMID23178123.
- ^Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M (March 2013)."Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products".Proceedings of the National Academy of Sciences of the United States of America.110(13): 5046–5051.Bibcode:2013PNAS..110.5046D.doi:10.1073/pnas.1221724110.PMC3612664.PMID23479637.
- ^Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, Brandman O, Frost A (January 2015)."Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains".Science.347(6217): 75–78.Bibcode:2015Sci...347...75S.doi:10.1126/science.1259724.PMC4451101.PMID25554787.
- ^Keeley, J.; Gutnikoff, R. (2 January 2015)."Ribosome studies turn up new mechanism of protein synthesis"(Press release).Howard Hughes Medical Institute.Archivedfrom the original on 12 January 2015.Retrieved16 January2015.
- ^Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002)."Membrane-bound Ribosomes Define the Rough ER".Molecular Biology of the Cell(4th ed.). New York: Garland Science.ISBN978-0-8153-4072-0.
- ^Kressler, Dieter; Hurt, Ed; Babler, Jochen (2009)."Driving ribosome assembly"(PDF).Biochimica et Biophysica Acta (BBA) - Molecular Cell Research.1803(6): 673–683.doi:10.1016/j.bbamcr.2009.10.009.PMID19879902.
- ^Dance, Amber (28 February 2023)."How did life begin? One key ingredient is coming into view - A Nobel-prizewinning scientist's team has taken a big step forward in its quest to reconstruct an early-Earth RNA capable of building proteins".Nature.615(7950): 22–25.doi:10.1038/d41586-023-00574-4.PMID36854922.
- ^abNoller HF (April 2012)."Evolution of protein synthesis from an RNA world".Cold Spring Harbor Perspectives in Biology.4(4): a003681.doi:10.1101/cshperspect.a003681.PMC3312679.PMID20610545.
- ^Dabbs ER (1986).Mutant studies on the prokaryotic ribosome.New York: Springer-Verlag.
- ^Noller HF, Hoffarth V, Zimniak L (June 1992). "Unusual resistance of peptidyl transferase to protein extraction procedures".Science.256(5062): 1416–9.Bibcode:1992Sci...256.1416N.doi:10.1126/science.1604315.PMID1604315.
- ^Nomura M, Mizushima S, Ozaki M, Traub P, Lowry CV (1969). "Structure and function of ribosomes and their molecular components".Cold Spring Harbor Symposia on Quantitative Biology.34:49–61.doi:10.1101/sqb.1969.034.01.009.PMID4909519.
- ^Krupkin M, Matzov D, Tang H, Metz M, Kalaora R, Belousoff MJ, Zimmerman E, Bashan A, Yonath A (Oct 2011)."A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome".Phil. Trans. R. Soc. B.366(1580): 2972–8.doi:10.1098/rstb.2011.0146.PMC3158926.PMID21930590.
- ^Bose T, Fridkin G, Davidovich C, Krupkin M, Dinger N, Falkovich AH, Peleg Y, Agmon I, Bashan A, Yonat A (Feb 2022)."Origin of life: protoribosome forms peptide bonds and links RNA and protein dominated worlds".Nucleic Acids Res.50(4): 1815–1828.doi:10.1093/nar/gkac052.PMC8886871.PMID35137169.
- ^abRoot-Bernstein M, Root-Bernstein R (February 2015)."The ribosome as a missing link in the evolution of life".Journal of Theoretical Biology.367:130–158.doi:10.1016/j.jtbi.2014.11.025.PMID25500179.
- ^Yarus M (2002). "Primordial genetics: phenotype of the ribocyte".Annual Review of Genetics.36:125–51.doi:10.1146/annurev.genet.36.031902.105056.PMID12429689.
- ^Forterre P, Krupovic M (2012). "The Origin of Virions and Virocells: The Escape Hypothesis Revisited".Viruses: Essential Agents of Life.pp. 43–60.doi:10.1007/978-94-007-4899-6_3.ISBN978-94-007-4898-9.
- ^Caetano-Anollés G, Seufferheld MJ (2013). "The coevolutionary roots of biochemistry and cellular organization challenge the RNA world paradigm".Journal of Molecular Microbiology and Biotechnology.23(1–2): 152–77.doi:10.1159/000346551.PMID23615203.S2CID41725226.
- ^Saladino R, Botta G, Pino S, Costanzo G, Di Mauro E (August 2012). "Genetics first or metabolism first? The formamide clue".Chemical Society Reviews.41(16): 5526–65.doi:10.1039/c2cs35066a.hdl:11573/494138.PMID22684046.
- ^Fox GE (September 2010)."Origin and Evolution of the Ribosome".Cold Spring Harb Perspect Biol.2(9): a003483.doi:10.1101/cshperspect.a003483.PMC2926754.PMID20534711.
- ^Fox GE (2016). "Origins and early evolution of the ribosome". In Hernández G, Jagus R (eds.).Evolution of the Protein Synthesis Machinery and Its Regulation.Switzerland: Springer, Cham. pp. 31–60.doi:10.1007/978-3-319-39468-8.ISBN978-3-319-39468-8.S2CID27493054.
- ^Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, Barna M (July 2017)."Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide".Molecular Cell.67(1). Elsevier BV: 71–83.e7.doi:10.1016/j.molcel.2017.05.021.PMC5548184.PMID28625553.
- ^Xue S, Barna M (May 2012)."Specialized ribosomes: a new frontier in gene regulation and organismal biology".Nature Reviews. Molecular Cell Biology.13(6). Springer Science and Business Media LLC: 355–369.doi:10.1038/nrm3359.PMC4039366.PMID22617470.
- ^Ferretti MB, Karbstein K (May 2019)."Does functional specialization of ribosomes really exist?".RNA.25(5). Cold Spring Harbor Laboratory: 521–538.doi:10.1261/rna.069823.118.PMC6467006.PMID30733326.
- ^Farley-Barnes KI, Ogawa LM,Baserga SJ(October 2019)."Ribosomopathies: Old Concepts, New Controversies".Trends in Genetics.35(10). Elsevier BV: 754–767.doi:10.1016/j.tig.2019.07.004.PMC6852887.PMID31376929.
- ^Mauro VP, Edelman GM (September 2002)."The ribosome filter hypothesis".Proceedings of the National Academy of Sciences of the United States of America.99(19): 12031–6.Bibcode:2002PNAS...9912031M.doi:10.1073/pnas.192442499.PMC129393.PMID12221294.
- ^Xue S, Barna M (May 2012)."Specialized ribosomes: a new frontier in gene regulation and organismal biology".Nature Reviews. Molecular Cell Biology.13(6): 355–69.doi:10.1038/nrm3359.PMC4039366.PMID22617470.
- ^Mathis AD, Naylor BC, Carson RH, Evans E, Harwell J, Knecht J, Hexem E, Peelor FF, Miller BF, Hamilton KL, Transtrum MK, Bikman BT, Price JC (February 2017)."Mechanisms of In Vivo Ribosome Maintenance Change in Response to Nutrient Signals".Molecular & Cellular Proteomics.16(2): 243–254.doi:10.1074/mcp.M116.063255.PMC5294211.PMID27932527.
- ^Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, et al. (May 2012)."Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae".Genetics.191(1). Genetics Society of America: 107–118.doi:10.1534/genetics.111.136549.PMC3338253.PMID22377630.
- ^Lee SW, Berger SJ, Martinović S, Pasa-Tolić L, Anderson GA, Shen Y, et al. (April 2002)."Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR".Proceedings of the National Academy of Sciences of the United States of America.99(9): 5942–5947.Bibcode:2002PNAS...99.5942L.doi:10.1073/pnas.082119899.PMC122881.PMID11983894.
- ^Carroll AJ, Heazlewood JL, Ito J, Millar AH (February 2008)."Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification".Molecular & Cellular Proteomics.7(2): 347–369.doi:10.1074/mcp.m700052-mcp200.PMID17934214.
- ^Odintsova TI, Müller EC, Ivanov AV, Egorov TA, Bienert R, Vladimirov SN, et al. (April 2003). "Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing".Journal of Protein Chemistry.22(3): 249–258.doi:10.1023/a:1025068419698.PMID12962325.S2CID10710245.
- ^Yu Y, Ji H, Doudna JA, Leary JA (June 2005)."Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes".Protein Science.14(6): 1438–1446.doi:10.1110/ps.041293005.PMC2253395.PMID15883184.
- ^Zeidan Q, Wang Z, De Maio A, Hart GW (June 2010)."O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins".Molecular Biology of the Cell.21(12): 1922–1936.doi:10.1091/mbc.e09-11-0941.PMC2883937.PMID20410138.
- ^Landry DM, Hertz MI, Thompson SR (December 2009)."RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs".Genes & Development.23(23): 2753–2764.doi:10.1101/gad.1832209.PMC2788332.PMID19952110.
- ^Decatur WA, Fournier MJ (July 2002). "rRNA modifications and ribosome function".Trends in Biochemical Sciences.27(7): 344–51.doi:10.1016/s0968-0004(02)02109-6.PMID12114023.
- ^Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP (November 2017). "Visualization of chemical modifications in the human 80S ribosome structure".Nature.551(7681): 472–477.Bibcode:2017Natur.551..472N.doi:10.1038/nature24482.PMID29143818.S2CID4465175.
- ^Guo H (August 2018). "Specialized ribosomes and the control of translation".Biochemical Society Transactions.46(4): 855–869.doi:10.1042/BST20160426.PMID29986937.S2CID51609077.
External links
edit- Lab computer simulates ribosome in motion
- Role of the Ribosome,Gwen V. Childs, copiedhere
- RibosomeinProteopedia—The free, collaborative 3D encyclopedia of proteins & other molecules
- Ribosomal proteins families in ExPASyArchived2011-04-30 at theWayback Machine
- Molecule of the MonthArchived2009-10-27 at theWayback Machine© RCSB Protein Data Bank:
- RibosomeArchived2010-11-14 at theWayback Machine
- Elongation FactorsArchived2011-03-16 at theWayback Machine
- Palade
- 3D electron microscopy structures of ribosomes at the EM Data Bank (EMDB)
- This article incorporatespublic domain materialfromScience Primer.NCBI.Archived fromthe originalon 2009-12-08.