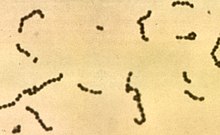Streptococcusis agenusofgram-positiveor spherical bacteria that belongs to the familyStreptococcaceae,within the orderLactobacillales(lactic acid bacteria), in the phylumBacillota.[2]Cell divisionin streptococci occurs along a singleaxis,thus when growing they tend to form pairs or chains, which may appear bent or twisted. This differs fromstaphylococci,which divide along multiple axes, thereby generating irregular, grape-like clusters ofcells.Most streptococci areoxidase-negativeandcatalase-negative,and many arefacultative anaerobes(capable of growth both aerobically and anaerobically).
The term was coined in 1877 by Viennese surgeonAlbert Theodor Billroth(1829–1894),[3]by combining the prefix "strepto-" (fromAncient Greek:στρεπτός,romanized:streptós,lit. 'easily twisted, pliant'[4]), together with the suffix "-coccus" (from ModernLatin:coccus,from Ancient Greek:κόκκος,romanized:kókkos,lit. 'grain, seed, berry'.[5]) In 1984, many bacteria formerly grouped in the genusStreptococcuswere separated out into thegeneraEnterococcusandLactococcus.[6]Currently, over 50 species are recognised in this genus. This genus has been found to be part of thesalivary microbiome.[7]
Pathogenesis and classification
editIn addition tostreptococcal pharyngitis(strep throat), certainStreptococcusspecies are responsible for many cases ofpink eye,[8]meningitis,bacterial pneumonia,endocarditis,erysipelas,andnecrotizing fasciitis(the 'flesh-eating' bacterial infections). However, many streptococcal species are not pathogenic, and form part of thecommensalhumanmicrobiotaof the mouth, skin, intestine, and upper respiratory tract. Streptococci are also a necessary ingredient in producingEmmentaler ( "Swiss" ) cheese.[9]
Species of streptococci are classified based on theirhemolyticproperties.[10]Alpha-hemolytic species cause oxidization of iron inhemoglobinmolecules within red blood cells, giving it a greenish color on blood agar.Beta-hemolyticspecies cause complete rupture of red blood cells. On blood agar, this appears as wide areas clear of blood cells surrounding bacterial colonies.Gamma-hemolyticspecies cause no hemolysis.[11]
Beta-hemolytic streptococci are further classified byLancefield grouping,aserotypeclassification (that is, describing specific carbohydrates present on the bacterial cell wall).[6]The 21 described serotypes are named Lancefield groups A to W (excluding E, I and J). This system of classification was developed byRebecca Lancefield,a scientist atRockefeller University.[12]
In the medical setting, the most important groups are the Alpha -hemolytic streptococciS. pneumoniaeandStreptococcusviridansgroups, and the beta-hemolytic streptococci of Lancefield groups A and B (also known as "group A strep" and "group B strep" ).
Table: Medically relevant streptococci[10]
| Species | Host | Disease |
|---|---|---|
| S. pyogenes | human | pharyngitis,cellulitis,erysipelas |
| S. agalactiae | human, cattle | neonatal meningitisandsepsis |
| S. dysgalactiae | human, animals | endocarditis,bacteremia,pneumonia,meningitis,respiratory infections |
| S. gallolyticus | human, animals | biliary orurinary tract infections,endocarditis |
| S. anginosus | human, animals | subcutaneous/organabscesses,meningitis,respiratory infections |
| S. sanguinis | human | endocarditis,dental caries |
| S. suis | swine | meningitis |
| S. mitis | human | endocarditis |
| S. mutans | human | dental caries |
| S. pneumoniae | human | pneumonia |
Alpha-hemolytic
editWhenAlpha -hemolysis(α-hemolysis) is present, the agar under the colony will appear dark and greenish due to the conversion of hemoglobin to greenbiliverdin.Streptococcus pneumoniaeand a group of oral streptococci (Streptococcus viridansor viridans streptococci) display Alpha -hemolysis. Alpha-hemolysis is also termed incomplete hemolysis or partial hemolysis because the cell membranes of the red blood cells are left intact. This is also sometimes called green hemolysis because of the color change in the agar.[citation needed]
Pneumococci
edit- S. pneumoniae(sometimes called pneumococcus), is a leading cause of bacterialpneumoniaand the occasional etiology ofotitis media,sinusitis,meningitis,andperitonitis.Inflammation is thought to be the major cause of how pneumococci cause disease, hence the tendency of diagnoses associated with them to involve inflammation. They possess no Lancefield antigens.[2]
The viridans group: Alpha -hemolytic
edit- Theviridans streptococciare a large group ofcommensalbacteria that are eitherAlpha -hemolytic,producing a green coloration on bloodagar plates(hence the name "viridans", from Latinvĭrĭdis,green), or nonhemolytic. They possess no Lancefield antigens.[2]
Beta-hemolytic
editBeta-hemolysis(β-hemolysis), sometimes called completehemolysis,is a complete lysis of red cells in the media around and under the colonies: the area appears lightened (yellow) and transparent. Streptolysin, an exotoxin, is the enzyme produced by the bacteria which causes the complete lysis of red blood cells. There are two types of streptolysin: Streptolysin O (SLO) and streptolysin S (SLS). Streptolysin O is an oxygen-sensitive cytotoxin, secreted by most group AStreptococcus(GAS), and interacts with cholesterol in the membrane of eukaryotic cells (mainly red and white blood cells, macrophages, and platelets), and usually results in beta-hemolysis under the surface of blood agar. Streptolysin S is an oxygen-stable cytotoxin also produced by most GAS strains which results in clearing on the surface of blood agar. SLS affects immune cells, including polymorphonuclear leukocytes and lymphocytes, and is thought to prevent the host immune system from clearing infection.Streptococcus pyogenes,or GAS, displays beta hemolysis.
Some weakly beta-hemolytic species cause intense hemolysis when grown together with a strain ofStaphylococcus.This is called theCAMP test.Streptococcus agalactiaedisplays this property.Clostridium perfringenscan be identified presumptively with this test.Listeria monocytogenesis also positive on sheep's blood agar.
Group A
editGroup AS. pyogenesis the causative agent in a wide range ofgroup A streptococcal infections(GAS). Theseinfectionsmay be noninvasive or invasive. The noninvasive infections tend to be more common and less severe. The most common of these infections includestreptococcal pharyngitis(strep throat) andimpetigo.[13]Scarlet feveris another example of Group A noninvasive infection.
The invasive infections caused by group A beta-hemolytic streptococci tend to be more severe and less common. This occurs when the bacterium is able to infect areas where it is not usually found, such as thebloodandorgans.[14]The diseases that may be caused include streptococcaltoxic shock syndrome,necrotizing fasciitis,pneumonia,andbacteremia.[13]Globally, GAS has been estimated to cause more than 500,000 deaths every year, making it one of the world's leadingpathogens.[13]
Additional complications may be caused by GAS, namely acuterheumatic feverand acuteglomerulonephritis.Rheumatic fever,a disease that affects thejoints,kidneys,andheart valves,is a consequence of untreated strep A infection caused not by the bacterium itself, but due to the antibodies created by the immune system to fight off the infection cross-reacting with other proteins in the body. This "cross-reaction" causes the body to essentially attack itself and leads to the damage above. A similar autoimmune mechanism initiated byGroup A beta-hemolytic streptococcal (GABHS) infectionis hypothesized to causepediatric autoimmune neuropsychiatric disorders associated with streptococcal infections (PANDAS),wherein autoimmune antibodies affect the basal ganglia, causing rapid onset of psychiatric, motor, sleep, and other symptoms in pediatric patients.
GAS infection is generally diagnosed with arapid strep testor by culture.
Group B
editS. agalactiae,or group Bstreptococcus,GBS,causes pneumonia and meningitis innewbornsand theelderly,with occasional systemicbacteremia.Importantly,Streptococcus agalactiaeis the most common cause of meningitis ininfantsfrom one month to three months old. They can also colonize the intestines and the female reproductive tract, increasing the risk for prematurerupture of membranesduring pregnancy, andtransmissionof the organism to the infant. TheAmerican College of Obstetricians and Gynecologists,American Academy of Pediatrics,and theCenters for Disease Controlrecommend all pregnant women between 35 and 37 weeks gestation to be tested for GBS. Women who test positive should be given prophylactic antibiotics during labor, which will usually prevent transmission to the infant.[15]Group III polysaccharide vaccines have been proven effective in preventing the passing of GBS from mother to infant.[16]
The United Kingdom has chosen to adopt a risk factor-based protocol, rather than the culture-based protocol followed in the US.[17]Current guidelines state that if one or more of the following risk factors is present, then the woman should be treated withintrapartumantibiotics:
- GBSbacteriuriaduring this pregnancy
- History of GBS disease in a previous infant
- Intrapartum fever (≥38 °C)
- Preterm labour (<37 weeks)
- Prolonged rupture of membranes (>18 hours)
This protocol results in the administration of intrapartum antibiotics to 15–20% of pregnant women and the prevention of 65–70% of cases of early onset GBS sepsis.[18]
Group C
editThis group includesS. equi,which causesstranglesin horses,[19]andS. zooepidemicus—S. equiis aclonaldescendant orbiovarof the ancestralS. zooepidemicus— which causes infections in several species of mammals, including cattle and horses.S. dysgalactiaesubsp. dysgalactiae[20]is also a member of group C,beta-haemolytic streptococcithat can causepharyngitisand otherpyogenicinfections similar togroup A streptococci.Group C streptococcal bacteria are considered zoonotic pathogens, meaning infection can be passed from animal to human.[21]
Group D (enterococci)
editMany former group D streptococci have been reclassified and placed in the genusEnterococcus(includingE. faecalis,E. faecium,E. durans,andE. avium).[22]For example,Streptococcus faecalisis nowEnterococcus faecalis.E. faecalisis sometimes Alpha -hemolytic andE. faeciumis sometimes beta hemolytic.[23]
The remaining nonenterococcal group D strains includeStreptococcus gallolyticus,Streptococcus bovis,Streptococcus equinusandStreptococcus suis.
Nonhemolytic streptococci rarely cause illness. However, weakly hemolytic group D beta-hemolytic streptococci andListeria monocytogenes(which is actually agram-positivebacillus) should not be confused with nonhemolytic streptococci.
Group F streptococci
editGroup F streptococci were first described in 1934 by Long andBlissamong the "minute haemolytic streptococci".[24]They are also known asStreptococcus anginosus(according to the Lancefield classification system) or as members of theS. millerigroup(according to the European system).
Group G streptococci
editThese streptococci are usually, but not exclusively, beta-hemolytic.Streptococcus dysgalactiaesubsp. canis[20]is the predominant subspecies encountered. It is a particularly common GGS in humans, although it is typically found on animals.S. phocaeis a GGS subspecies that has been found in marine mammals and marine fish species. In marine mammals it has been mainly associated withmeningoencephalitis,sepsis,andendocarditis,but is also associated with many other pathologies. Its environmental reservoir and means of transmission in marine mammals is not well characterized. Group G streptococci are also considered zoonotic pathogens.
Group H streptococci
editGroup H streptococci cause infections in medium-sized canines. Group H streptococci rarely cause human illness unless a human has direct contact with the mouth of a canine. One of the most common ways this can be spread is human-to-canine, mouth-to-mouth contact. However, the canine may lick the human's hand and infection can be spread, as well.[25]
Clinical identification
editIn clinical practice, the most common groups ofStreptococcuscan be distinguished by simple bench tests, such as the PYR test forgroup A streptococcus.There are also latex agglutination kits which can distinguish each of the main groups seen in clinical practice.
Treatment
editStreptococcal infections can be treated with antibiotics from thepenicillinfamily. Most commonly, penicillin or amoxicillin is used to treat strep infection. These antibiotics work by disrupting peptidoglycan production in the cell wall.[26]Treatment most often occurs as a 10-day oral antibiotic cycle. For patients with penicillin allergies and those suffering from skin infections, clindamycin can be used. Clindamycin works by disrupting protein synthesis within the cell.
Molecular taxonomy and phylogenetics
editStreptococci have been divided into six groups on the basis of their16SrDNA sequences:S. anginosus, S. gallolyticus, S. mitis, S. mutans, S. pyogenesandS. salivarius.[28]The 16S groups have been confirmed by whole genome sequencing (see figure). The important pathogensS. pneumoniaeandS. pyogenesbelong to theS. mitisandS. pyogenesgroups, respectively,[29]while the causative agent ofdental caries,Streptococcus mutans,is basal to theStreptococcusgroup.
Recent technological advances have resulted in an increase of available genome sequences forStreptococcusspecies, allowing for more robust and reliable phylogenetic and comparative genomic analyses to be conducted.[30]In 2018, the evolutionary relationships withinStreptococcuswas re-examined by Patel and Gupta through the analysis of comprehensivephylogenetic treesconstructed based on four different datasets of proteins and the identification of 134 highly specific molecular signatures (in the form ofconserved signature indels) that are exclusively shared by the entire genus or its distinct subclades.[30]
The results revealed the presence of two main clades at the highest level withinStreptococcus,termed the "Mitis-Suis" and "Pyogenes-Equinus-Mutans" clades.[30]The "Mitis-Suis" main clade comprises the Suis subclade and the Mitis clade, which encompasses the Angiosus, Pneumoniae, Gordonii and Parasanguinis subclades. The second main clade, the "Pyogenes-Equinus-Mutans", includes the Pyogenes, Mutans, Salivarius, Equinus, Sobrinus, Halotolerans, Porci, Entericus and Orisratti subclades. In total, 14 distinct subclades have been identified within the genusStreptococcus,each supported by reliable branching patterns in phylogenetic trees and by the presence of multipleconserved signature indelsin different proteins that are distinctive characteristics of the members of these 14 clades.[30]A summary diagram showing the overall relationships among theStreptococcusbased on these studies is depicted in a figure on this page.
Genomics
editThe genomes of hundreds of species have been sequenced.[32]MostStreptococcusgenomes are 1.8 to 2.3 Mb in size and encode 1,700 to 2,300 proteins. Some important genomes are listed in the table.[33]The four species shown in the table (S. pyogenes, S. agalactiae, S. pneumoniae,andS. mutans) have an average pairwise protein sequence identity of about 70%.[33]
| feature | S. pyogenes | S. agalactiae | S. pneumoniae | S. mutans |
|---|---|---|---|---|
| base pairs | 1,852,442 | 2,211,488 | 2,160,837 | 2,030,921 |
| ORFs | 1792 | 2118 | 2236 | 1963 |
| prophages | yes | no | no | no |
Bacteriophage
editBacteriophageshave been described for many species ofStreptococcus.18prophageshave been described inS. pneumoniaethat range in size from 38 to 41 kb in size, encoding from 42 to 66 genes each.[34]Some of the firstStreptococcusphages discovered were Dp-1[35][36] and ω1 (alias ω-1).[37][38][39] In 1981 the Cp (Complutense phage 1, officiallyStreptococcus virus Cp1,Picovirinae) family was discovered with Cp-1 as its first member.[40]Dp-1 and Cp-1 infect bothS. pneumoniaeandS. mitis.[41]However, the host ranges of mostStreptococcusphages have not been investigated systematically.
Natural genetic transformation
editNatural genetic transformationinvolves the transfer of DNA from one bacterium to another through the surrounding medium. Transformation is a complex process dependent on the expression of numerous genes. To be capable of transformation a bacterium must enter a special physiologic state referred to ascompetence.S. pneumoniae,S. mitisandS. oraliscan become competent, and as a result actively acquire homologous DNA for transformation by a predatory fratricidal mechanism[42]This fratricidal mechanism mainly exploits non-competent siblings present in the same niche[43]Among highly competent isolates ofS. pneumoniae,Li et al.[44]showed that nasal colonization fitness and virulence (lung infectivity) depend on an intact competence system. Competence may allow the streptococcal pathogen to use external homologous DNA for recombinational repair of DNA damages caused by the host's oxidative attack.[45]
See also
editReferences
edit- ^abcdefghijklmnopqrstuvwxyzaaabacadaeafagahaiajakalamanaoapaqarasatauavawaxayazbabbbcbdbebfbgbhbibjbkblbmbnbobpbqbrbsbtParte AC."Streptococcus".LPSN.
- ^abcRyan KJ, Sherris JC, eds. (1994).Sherris Medical Microbiology(3rd ed.). Appleton & Lange. pp.266–7.ISBN0-8385-8541-8.
- ^"streptococcus".Online Etymology Dictionary.Retrieved25 July2018.
- ^στρεπτόςinLiddell, Henry George;Scott, Robert(1940)A Greek–English Lexicon,revised and augmented throughout byJones, Sir Henry Stuart,with the assistance of McKenzie, Roderick. Oxford: Clarendon Press. In thePerseus Digital Library,Tufts University.
- ^κόκκοςinLiddellandScott
- ^abFacklam R (October 2002)."What happened to the streptococci: overview of taxonomic and nomenclature changes".Clinical Microbiology Reviews.15(4): 613–630.doi:10.1128/CMR.15.4.613-630.2002.PMC126867.PMID12364372.
- ^Wang K, Lu W, Tu Q, Ge Y, He J, Zhou Y, et al. (March 2016)."Preliminary analysis of salivary microbiome and their potential roles in oral lichen planus".Scientific Reports.6(1): 22943.Bibcode:2016NatSR...622943W.doi:10.1038/srep22943.PMC4785528.PMID26961389.
- ^"How to Get Rid of Pinkeye, Symptoms, Treatment, Causes & Pictures".
- ^"Streptococcus | Center for Academic Research and Training in Anthropogeny (CARTA)".carta.anthropogeny.org.Retrieved2022-07-23.
- ^abPatterson MJ (1996). Baron S; et al. (eds.).Streptococcus.In:Baron's Medical Microbiology(4th ed.). Univ of Texas Medical Branch.ISBN978-0-9631172-1-2.(via NCBI Bookshelf).
- ^Sharma S, Khanna G, Gangane SD (2019-07-13).Textbook of Pathology and Genetics for Nurses E-Book.Elsevier Health Sciences.ISBN978-81-312-5538-4.
- ^Carroll KC (August 2019). Munson E (ed.)."Biographical Feature: Rebecca Lancefield, Ph.D".Journal of Clinical Microbiology.57(8).doi:10.1128/JCM.00728-19.PMC6663886.PMID31142605.
- ^abcCohen-Poradosu R, Kasper DL (October 2007)."Group A streptococcus epidemiology and vaccine implications".Clinical Infectious Diseases.45(7): 863–865.doi:10.1086/521263.PMID17806050.
- ^"Streptococcal Infections (Invasive Group A Strep)".New York City Department of Health and Mental Hygiene. Archived fromthe originalon 6 November 2012.Retrieved21 November2012.
- ^Schrag S, Gorwitz R, Fultz-Butts K, Schuchat A (August 2002). "Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC".MMWR. Recommendations and Reports.51(RR-11): 1–22.PMID12211284.
- ^Noya, Francisco J. D.; Baker, Carol J. (1992-03-01)."PREVENTION OF GROUP B STREPTOCOCCAL INFECTION".Infectious Disease Clinics of North America.6(1): 41–55.doi:10.1016/S0891-5520(20)30424-4.ISSN0891-5520.PMID1578122.
- ^"Prevention of Early-onset Neonatal Group B Streptococcal Disease: Green-top Guideline No. 36".BJOG.124(12): e280–e305. November 2017.doi:10.1111/1471-0528.14821.PMID28901693.
- ^Norwitz ER, Schorge JO (2013).Obstetrics and Gynecology at a Glance(4th ed.). Chichester: John Wiley & Sons, Ltd.ISBN978-1118341735.
- ^Harrington DJ, Sutcliffe IC, Chanter N (April 2002)."The molecular basis of Streptococcus equi infection and disease".Microbes and Infection.4(4): 501–510.doi:10.1016/S1286-4579(02)01565-4.PMID11932201.
- ^abHaslam DB, St Geme III JW (2023). "122 - Groups C and G Streptococci". In Long SS, Prober CG, Fischer M, Kimberlin D (eds.).Principles and Practice of Pediatric Infectious Diseases(Sixth ed.). Elsevier. pp. 752–753.doi:10.1016/B978-0-323-75608-2.00122-1.ISBN978-0-323-75608-2.Note that according to the same source, the subspeciesequisimilisis a grouping of largeS. dysgalactiaecolonies, whether they are members of Group C or Group G.
- ^Klos, Marta (12 June 2017)."Pathogenicity of Virulent Species of Group C Streptococci in Human".Can J Infect Dis Med Microbiol.2017:1–5.doi:10.1155/2017/9509604.PMC5485279.PMID28694832.
- ^Köhler W (June 2007). "The present state of species within the genera Streptococcus and Enterococcus".International Journal of Medical Microbiology.297(3): 133–150.doi:10.1016/j.ijmm.2006.11.008.PMID17400023.
- ^Holt et al. (1994).Bergey's Manual of Determinative Bacteriology(9th ed.). Lippincott Williams & Wilkins.ISBN0-683-00603-7
- ^Whitworth JM (November 1990)."Lancefield group F and related streptococci".Journal of Medical Microbiology.33(3): 135–151.doi:10.1099/00222615-33-3-135.PMID2250284.[permanent dead link]
- ^"Bacterial Infection (Streptococcus) in Dogs".petmd.Retrieved12 December2014.
- ^Lowe, Derek (19 Jan 2022)."How Do Penicillins Actually Work?".Science– via American Association for the Advancement of Science.
- ^"Bacteria-Firmicutes-Bacilli-Lactobacillales-Streptococcaceae-Streptococcus".PATRIC, University of Chicago.Retrieved12 December2014.
- ^Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T (April 1995)."Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus".International Journal of Systematic Bacteriology.45(2): 406–408.doi:10.1099/00207713-45-2-406.PMID7537076.
- ^Liu, D.,Molecular Detection of Human Bacterial Pathogens(Boca Raton:CRC Press,2011),p. 324.
- ^abcdePatel S, Gupta RS (December 2018). "Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures".Infection, Genetics and Evolution.66:130–151.Bibcode:2018InfGE..66..130P.doi:10.1016/j.meegid.2018.09.020.PMID30248475.S2CID52813184.
- ^Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, et al. (April 2007)."Genome of the opportunistic pathogen Streptococcus sanguinis".Journal of Bacteriology.189(8): 3166–3175.doi:10.1128/JB.01808-06.PMC1855836.PMID17277061.
- ^"Streptococcus".PATRIC.Blacksburg, VA: Virginia Bioinformatics Institute. Archived fromthe originalon 2013-03-10.
- ^abFerretti JJ, Ajdic D, McShan WM (May 2004). "Comparative genomics of streptococcal species".The Indian Journal of Medical Research.119(Suppl): 1–6.PMID15232152.
- ^McShan, W. Michael; Nguyen, Scott V. (2016), Ferretti, Joseph J.; Stevens, Dennis L.; Fischetti, Vincent A. (eds.),"The Bacteriophages of Streptococcus pyogenes",Streptococcus pyogenes: Basic Biology to Clinical Manifestations,Oklahoma City (OK): University of Oklahoma Health Sciences Center,PMID26866212,retrieved2024-02-07
- ^ McDonnell M, Ronda C, Tomasz A (1975) "Diplophage": a bacteriophage of Diplococcus pneumoniae. Virology 63:577–582
- ^NCBI:Streptococcus phage Dp-1(species)
- ^Tiraby JG, Tiraby E, Fox MS (Dec 1975) Pneumococcal bacteriophages. Virology 68:566–569.doi:10.1016/0042-6822(75)90300-1.PMID844
- ^López R (September 2004). "Streptococcus pneumoniae and its bacteriophages: one long argument".International Microbiology.7(3): 163–171.PMID15492930.PDF via web archive(9 Aug 2017)
- ^Rubens López, Ernesto García:Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage,Oxford Academic FEMS Microbiology Reviews. Volume 28, Issue 5. 1 Nov 2004, pp. 554—580,doi:10.1016/j.femsre.2004.05.002(Free Fulltext)
- ^Ronda C, López R, García E (November 1981)."Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae".Journal of Virology.40(2): 551–559.doi:10.1128/JVI.40.2.551-559.1981.PMC256658.PMID6275103.
- ^Ouennane S, Leprohon P, Moineau S (2015)."Diverse virulent pneumophages infect Streptococcus mitis".PLOS ONE.10(2): e0118807.Bibcode:2015PLoSO..1018807O.doi:10.1371/journal.pone.0118807.PMC4334900.PMID25692983.
- ^Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS (July 2008)."A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species".Molecular Microbiology.69(1): 245–253.doi:10.1111/j.1365-2958.2008.06288.x.PMID18485065.S2CID30923996.
- ^Claverys JP, Håvarstein LS (March 2007). "Cannibalism and fratricide: mechanisms and raisons d'être".Nature Reviews. Microbiology.5(3): 219–229.doi:10.1038/nrmicro1613.PMID17277796.S2CID35433490.
- ^Li G, Liang Z, Wang X, Yang Y, Shao Z, Li M, et al. (June 2016)."Addiction of Hypertransformable Pneumococcal Isolates to Natural Transformation for In Vivo Fitness and Virulence".Infection and Immunity.84(6): 1887–1901.doi:10.1128/IAI.00097-16.PMC4907133.PMID27068094.
- ^Michod RE, Bernstein H, Nedelcu AM (May 2008). "Adaptive value of sex in microbial pathogens".Infection, Genetics and Evolution.8(3): 267–285.Bibcode:2008InfGE...8..267M.doi:10.1016/j.meegid.2008.01.002.PMID18295550.
External links
edit- Centers for Disease Control and Prevention (CDC) (March 2000)."Adoption of perinatal group B streptococcal disease prevention recommendations by prenatal-care providers--Connecticut and Minnesota, 1998".MMWR. Morbidity and Mortality Weekly Report.49(11): 228–232.PMID10763673.
- Nature-Inspired CRISPR Enzyme Discoveries Vastly Expand Genome Editing.On: SciTechDaily. June 16, 2020. Source: Media Lab, Massachusetts Institute of Technology.
- Streptococcusgenomes and related information atPATRIC,a Bioinformatics Resource Center funded byNIAID
- The Canadian Strep B FoundationArchived2013-05-02 at theWayback Machine
- The UK Group B Strep Supportcharity
- StutteringStreptococcal InfectionInfection