Negative-strand RNA virus
| Negarnaviricota | |
|---|---|

| |
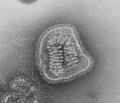
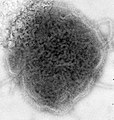
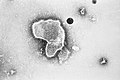
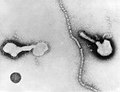
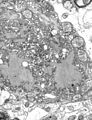
| A montage oftransmission electron micrographsof some viruses in the phylumNegarnaviricota.Not to scale. Species from left to right, top to bottom:Zaire ebolavirus,Sin Nombre orthohantavirus,Human orthopneumovirus,Hendra henipavirus,an unidentifiedrhabdovirus,Measles morbillivirus. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Subtaxa | |
| Synonyms[1][2] | |
| |
Negative-strand RNA viruses(−ssRNA viruses) are a group of relatedvirusesthat havenegative-sense,single-stranded genomes made ofribonucleic acid(RNA). They have genomes that act as complementary strands from whichmessenger RNA(mRNA) is synthesized by the viral enzymeRNA-dependent RNA polymerase(RdRp). During replication of the viral genome, RdRp synthesizes a positive-sense antigenome that it uses as a template to create genomic negative-sense RNA. Negative-strand RNA viruses also share a number of other characteristics: most contain aviral envelopethat surrounds the capsid, which encases the viral genome, −ssRNA virus genomes are usually linear, and it is common for their genome to be segmented.
Negative-strand RNA viruses constitute thephylumNegarnaviricota,in the kingdomOrthornaviraeand realmRiboviria.They are descended from a common ancestor that was adouble-stranded RNA (dsRNA) virus,and they are considered to be a sister clade ofreoviruses,which are dsRNA viruses. Within the phylum, there are two major branches that form two subphyla:Haploviricotina,whose members are mostly non-segmented and which encode an RdRp that synthesizes caps on mRNA, andPolyploviricotina,whose members are segmented and which encode an RdRp thatsnatches capsfrom host mRNAs. A total of six classes in the phylum are recognized.
Negative-strand RNA viruses are closely associated witharthropodsand can be informally divided between those that are reliant on arthropods for transmission and those that are descended from arthropod viruses but can now replicate in vertebrates without the aid of arthropods. Prominent arthropod-borne −ssRNA viruses include theRift Valley fever virusand thetomato spotted wilt virus.Notable vertebrate −ssRNA viruses include theEbola virus,hantaviruses,influenza viruses,theLassa fever virus,and therabies virus.
Etymology
[edit]Negarnaviricotatakes the first part of its name fromLatinnega,meaning negative, the middle partrnarefers to RNA, and the final part,viricota,is the suffix used for virus phyla. The subphylumHaploviricotinatakes the first part of its name,Haplo,fromAncient Greekἁπλός, meaning simple, and 'viricotinais the suffix used for virus subphyla. The subphylumPolyploviricotinafollows the same pattern,Polyplobeing taken from Ancient Greek πολύπλοκος, meaning complex.[1]
Characteristics
[edit]Genome
[edit]
All viruses inNegarnaviricotaare negative-sense, single-stranded RNA (−ssRNA) viruses. They have genomes made of RNA, which are single instead of double-stranded. Their genomes are negative sense, meaning that messenger RNA (mRNA) can be synthesized directly from the genome by the viral enzyme RNA-dependent RNA polymerase (RdRp), also called RNA replicase, which is encoded by all −ssRNA viruses. Excluding viruses in the genusTenuivirusand some in the familyChuviridae,all −ssRNA viruses have linear rather than circular genomes, and the genomes may be segmented or non-segmented.[1][3][4]All −ssRNA genomes containterminal inverted repeats,which are palindromic nucleotide sequences at each end of the genome.[5]
Replication and transcription
[edit]
Replication of −ssRNA genomes is executed by RdRp, which initiates replication by binding to a leader sequence on the 3'-end (usually pronounced "three prime end" ) of the genome. RdRp then uses the negative sense genome as a template to synthesize a positive-sense antigenome. When replicating the antigenome, RdRp first binds to the trailer sequence on the 3'-end of the antigenome. Thereafter, RdRp ignores all transcription signals on the antigenome and synthesizes a copy of the genome while using the antigenome as a template.[6]Replication is executed while the genome is inside the nucleocapsid, and RdRp unveils the capsid and translocates along the genome during replication. As new nucleotide sequences are synthesized by RdRp, capsid proteins are assembled and encapsidate the newly replicate viral RNA.[2]
Transcribing mRNA from the genome follows the same directional pattern as producing the antigenome. At the leader sequence, RdRp synthesizes a 5-'end (usually pronounced "five prime end" ) triphosphate-leader RNA and either, in the case of the subphylumHaploviricotina,caps the 5'-end or, in the case of the subphylumPolyploviricotina,snatches a capfrom a host mRNA and attaches it to the viral mRNA so that the mRNA can betranslatedby the host cell'sribosomes.[7][8][9]
After capping the mRNA, RdRp initiates transcription at a gene start signal and later terminates transcription upon reaching a gene end signal.[10]At the end of transcription, RdRp synthesizes apolyadenylated tail(poly (A) tail) consisting of hundreds ofadeninesin the mRNA's 3-end, which may be done bystutteringon a sequence ofuracils.[11][12]After the poly (A) tail is constructed, the mRNA is released by RdRp. In genomes that encode more than one transcribable portion, RdRp can continue scanning to the next start sequence to continue with transcription.[10][7][13]
Some −ssRNA viruses areambisense,meaning that both the negative genomic strand and positive antigenome separately encode different proteins. In order to transcribe ambisense viruses, two rounds of transcription are performed: first, mRNA is produced directly from the genome; second, mRNA is created from the antigenome. All ambisense viruses contain ahairpin loopstructure to stop transcription after the protein's mRNA has been transcribed.[14]
Morphology
[edit]
Negative-strand RNA viruses contain aribonucleoproteincomplex composed of the genome and an RdRp attached to each segment of the genome surrounded by a capsid.[15]The capsid is composed of proteins whose folded structure contains five alpha-helices in theN-terminallobe (5-H motif) and three alpha-helices in theC-terminallobe (3-H motif). Inside the capsid, the genome is sandwiched between these two motifs.[2]Excluding the familyAspiviridae,−ssRNA viruses contain an outerviral envelope,a type of alipidmembrane that surrounds the capsid. The shape of the virus particle, called a virion, of −ssRNA viruses varies and may be filamentous, pleomorphic, spherical, or tubular.[16]
Evolution
[edit]Genome segmentation is a prominent trait among many −ssRNA viruses, and −ssRNA viruses range from having genomes with one segment, typical for members of the orderMononegavirales,to genomes with ten segments, as is the case forTilapia tilapinevirus.[5][17]There is no clear trend over time that determines the number of segments, and genome segmentation among −ssRNA viruses appears to be a flexible trait since it has evolved independently on multiple occasions. Most members of the subphylumHaploviricotinaare nonsegmented, whereas segmentation is universal inPolyploviricotina.[2][5]
Phylogenetics
[edit]
Phylogenetic analysis based on RdRp shows that −ssRNA viruses are descended from a common ancestor and that they are likely a sister clade ofreoviruses,which are dsRNA viruses. Within the phylum, there are two clear branches, assigned to two subphyla, based on whether RdRp synthesizes a cap on viral mRNA orsnatches a capfrom host mRNA and attaches that cap to viral mRNA.[1][3]
Within the phylum, −ssRNA viruses that infect arthropods appear to be basal and the ancestors of all other −ssRNA viruses. Arthropods frequently live together in large groups, which allows for viruses to be transmitted easily. Over time, this has led to arthropod −ssRNA viruses gaining a high level of diversity. While arthropods host large quantities of viruses, there is disagreement about the degree to which cross-species transmission of arthropod −ssRNA viruses occurs among arthropods.[4][5]
Plant and vertebrate −ssRNA viruses tend to be genetically related to arthropod-infected viruses. Furthermore, most −ssRNA viruses outside of arthropods are found in species that interact with arthropods. Arthropods therefore serve as both key hosts and vectors of transmission of −ssRNA viruses. In terms of transmission, non-arthropod −ssRNA viruses can be distinguished between those that are reliant on arthropods for transmission and those that can circulate among vertebrates without the aid of arthropods. The latter group is likely to have originated from the former, adapting to vertebrate-only transmission.[5]
Classification
[edit]Negarnaviricotabelongs to the kingdomOrthornavirae,which encompasses all RNA viruses that encode RdRp, and the realmRiboviria,which includesOrthornaviraeas well as all viruses that encodereverse transcriptasein the kingdomPararnavirae.Negarnaviricotacontains two subphyla, which contain a combined six classes, five of which are monotypic down to lower taxa:[2][9][18]
- Subphylum:Haploviricotina,which contains −ssRNA viruses that encode an RdRp that synthesizes a cap structure on viral mRNA and which usually have nonsegmented genomes
- Class:Chunquiviricetes
- Order:Muvirales
- Family:Qinviridae
- Genus:Yingvirus
- Family:Qinviridae
- Order:Muvirales
- Class:Milneviricetes
- Order:Serpentovirales
- Family:Aspviridae
- Genus:Ophiovirus
- Family:Aspviridae
- Order:Serpentovirales
- Class:Monjiviricetes
- Class:Yunchangviricetes
- Order:Goujianvirales
- Family:Yueviridae
- Genus:Yuyuevirus
- Family:Yueviridae
- Order:Goujianvirales
- Class:Chunquiviricetes
- Subphylum:Polyploviricotina,which contains −ssRNA viruses that encode an RdRp that takes a cap from host mRNA to use as the cap on viral mRNA and which have segmented genomes
- Class:Ellioviricetes
- Order:Bunyavirales
- Class:Insthoviricetes
- Order:Articulavirales
- Class:Ellioviricetes
Negative-strand RNA viruses are classified as Group V in theBaltimore classificationsystem, which groups viruses together based on their manner of mRNA production and which is often used alongside standard virus taxonomy, which is based on evolutionary history. Therefore, Group V andNegarnaviricotaare synonymous.[1]
Disease
[edit]Negative-strand RNA viruses caused many widely known diseases. Many of these are transmitted by arthropods, including theRift Valley fever virusand thetomato spotted wilt virus.[19][20]Among vertebrates, bats and rodents are common vectors for many viruses, including theEbola virusand therabies virus,transmitted by bats and other vertebrates,[21][22]and theLassa fever virusandhantaviruses,transmitted by rodents.[23][24]Influenza virusesare common among birds and mammals.[25]Human-specific −ssRNA viruses include the measles virus and themumps virus.[26][27]
History
[edit]Many diseases caused by −ssRNA viruses have been known throughout history, including hantavirus infection, measles, and rabies.[28][29][30]In modern history, some such asEbolaandinfluenzahave caused deadly disease outbreaks.[31][32]Thevesicular stomatitis virus,first isolated in 1925 and one of the first animal viruses to be studied because it could be studied well incell cultures,was identified as an −ssRNA virus, which was unique at the time because other RNA viruses that had been discovered were positive sense.[33][34]In the early 21st century, the bovine diseaserinderpest,caused by −ssRNA rinderpest virus, became the second disease to be eradicated, aftersmallpox,caused by a DNA virus.[35]
In the 21st century,viral metagenomicshas become common to identify viruses in the environment. For −ssRNA viruses, this allowed for a large number of invertebrate, and especially arthropod, viruses to be identified, which helped to provide insight into the evolutionary history of −ssRNA viruses. Based on phylogenetic analysis of RdRp showing that −ssRNA viruses were descended from a common ancestor,Negarnaviricotaand its two subphyla were established in 2018, and it was placed into the then newly established realmRiboviria.[1][36]
Gallery
[edit]Notes
[edit]- ^Thehepatitis D virusis often called a virus but can be more specifically described as avirusoid-like pathogenic −ssRNA strand. It is excluded fromNegarnaviricotabecause although it is −ssRNA, it does not encode RdRp, which is the unifying trait of viruses inOrthornavirae.
References
[edit]- ^abcdefWolf Y, Krupovic M, Zhang YZ, Maes P, Dolji V, Koonin EV (21 August 2017)."Megataxonomy of negative-sense RNA viruses"(docx).International Committee on Taxonomy of Viruses (ICTV).Retrieved6 August2020.
- ^abcdeLuo M, Terrel JR, Mcmanus SA (30 July 2020)."Nucleocapsid Structure of Negative Strand RNA Virus".Viruses.12(8): 835.doi:10.3390/v12080835.PMC7472042.PMID32751700.
- ^abWolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Kooning EV (27 November 2018)."Origins and Evolution of the Global RNA Virome".mBio.9(6): e02329-18.doi:10.1128/mBio.02329-18.PMC6282212.PMID30482837.
- ^abKäfer S, Paraskevopoulou S, Zirkel F, Wieseke N, Donath A, Petersen M, Jones TC, Liu S, Zhou X, Middendorf M, Junglen S, Misof B, Drosten C (12 December 2019)."Re-assessing the diversity of negative strand RNA viruses in insects".PLOS Pathog.15(12): e1008224.doi:10.1371/journal.ppat.1008224.PMC6932829.PMID31830128.
- ^abcdeLi CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ (29 January 2015)."Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses".eLife.4(4): e05378.doi:10.7554/eLife.05378.PMC4384744.PMID25633976.
- ^"Negative stranded RNA virus replication".ViralZone.Swiss Institute of Bioinformatics.Retrieved6 August2020.
- ^ab"Negative-stranded RNA virus transcription".ViralZone.Swiss Institute of Bioinformatics.Retrieved6 August2020.
- ^"Cap snatching".ViralZone.Swiss Institute of Bioinformatics.Retrieved6 August2020.
- ^abKuhn JH, Wolf YI, Krupovic M, Zhang YZ, Maes P, Dolja VV, Koonin EV (February 2019)."Classify viruses - the gain is worth the pain".Nature.566(7744): 318–320.Bibcode:2019Natur.566..318K.doi:10.1038/d41586-019-00599-8.PMC11165669.PMID30787460.S2CID67769904.Retrieved6 August2020.
- ^abFearns, Rachel; Plemper, Richard K (15 April 2017)."Polymerases of paramyxoviruses and pneumoviruses".Virus Research.234:87–102.doi:10.1016/j.virusres.2017.01.008.ISSN0168-1702.PMC5476513.PMID28104450.
- ^Harmon, Shawn B.; Megaw, A. George; Wertz, Gail W. (1 January 2001)."RNA Sequences Involved in Transcriptional Termination of Respiratory Syncytial Virus".Journal of Virology.75(1): 36–44.doi:10.1128/JVI.75.1.36-44.2001.PMC113895.PMID11119571.
- ^Jacques, J. P.; Kolakofsky, D. (1 May 1991)."Pseudo-templated transcription in prokaryotic and eukaryotic organisms".Genes & Development.5(5): 707–713.doi:10.1101/gad.5.5.707.ISSN0890-9369.PMID2026325.
- ^"Negative-stranded RNA virus polymerase stuttering".ViralZone.Swiss Institute of Bioinformatics.Retrieved6 August2020.
- ^"Ambisense transcription in negative stranded RNA viruses".ViralZone.Swiss Institute of Bioinformatics.Retrieved6 August2020.
- ^Zhou H, Sun Y, Guo Y, Lou Z (September 2013). "Structural perspective on the formation of ribonucleoprotein complex in negative-sense single-stranded RNA viruses".Trends Microbiol.21(9): 475–484.doi:10.1016/j.tim.2013.07.006.PMID23953596.
- ^Fermin, G. (2018).Viruses: Molecular Biology, Host Interactions and Applications to Biotechnology.Elsevier. pp. 19–27, 43.doi:10.1016/B978-0-12-811257-1.00002-4.ISBN9780128112571.S2CID89706800.
- ^Bacharach E, Mishra N, Briese T, Zody MC, Kembou Tsofack JE, Zamostiano R, Berkowitz A, Ng J, Nitido A, Corvelo A, Toussaint NC, Abel Nielsen SC, Hornig M, Del Pozo J, Bloom T, Ferguson H, Eldar A, Lipkin WI (5 April 2016)."Characterization of a Novel Orthomyxo-like Virus Causing Mass Die-Offs of Tilapia".mBio.7(2): e00431-16.doi:10.1128/mBio.00431-16.PMC4959514.PMID27048802.
- ^"Virus Taxonomy: 2019 Release".talk.ictvonline.org.International Committee on Taxonomy of Viruses.Retrieved6 August2020.
- ^Hartman A (June 2017)."Rift Valley Fever".Clin Lab Med.37(2): 285–301.doi:10.1016/j.cll.2017.01.004.PMC5458783.PMID28457351.
- ^Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C, Foster GD (December 2011)."Top 10 plant viruses in molecular plant pathology".Mol Plant Pathol.12(9): 938–954.doi:10.1111/j.1364-3703.2011.00752.x.PMC6640423.PMID22017770.
- ^Muñoz-Fontela C, McElroy AK (2017)."Ebola Virus Disease in Humans: Pathophysiology and Immunity".Curr Top Microbiol Immunol.Current Topics in Microbiology and Immunology.411:141–169.doi:10.1007/82_2017_11.ISBN978-3-319-68946-3.PMC7122202.PMID28653186.
- ^Fisher CR, Streicker DG, Schnell MJ (April 2018)."The spread and evolution of rabies virus: conquering new frontiers".Nat Rev Microbiol.16(4): 241–255.doi:10.1038/nrmicro.2018.11.PMC6899062.PMID29479072.
- ^Yun NE, Walker DH (9 October 2012)."Pathogenesis of Lassa fever".Viruses.4(10): 2031–2048.doi:10.3390/v4102031.PMC3497040.PMID23202452.
- ^Avsic-Zupanc T, Saksida A, Korva M (April 2019)."Hantavirus infections".Clin Microbiol Infect.21S:e6–e16.doi:10.1111/1469-0691.12291.PMID24750436.Retrieved6 August2020.
- ^Borkenhagen LK, Salman MD, Ma MJ, Gray GC (November 2019)."Animal influenza virus infections in humans: A commentary".Int J Infect Dis.88:113–119.doi:10.1016/j.ijid.2019.08.002.PMID31401200.Retrieved6 August2020.
- ^"Transmission of Measles".cdc.gov.Centers for Disease Control and Prevention (CDC). 5 February 2018.Retrieved6 August2020.
- ^Rubin S, Eckhaus M, Rennick LJ, Bamford CG,Duprex WP(January 2015)."Molecular biology, pathogenesis and pathology of mumps virus".J Pathol.235(2): 242–252.doi:10.1002/path.4445.PMC4268314.PMID25229387.
- ^Jiang H, Zheng X, Wang L, Du H, Wang P, Bai X (2017)."Hantavirus infection: a global zoonotic challenge".Virol Sin.32(1): 32–43.doi:10.1007/s12250-016-3899-x.PMC6598904.PMID28120221.
- ^"Measles history".cdc.gov.Centers for Disease Control and Prevention (CDC). 5 February 2018.Retrieved6 August2020.
- ^Velasco-Villa A, Mauldin MR, Shi M, Escobar LE, Gallardo-Romero NF, Damon I, Olson VA, Streicker DG, Emerson G (October 2017)."The history of rabies in the Western Hemisphere".Antiviral Res.146:221–232.doi:10.1016/j.antiviral.2017.03.013.PMC5620125.PMID28365457.
- ^Zawilinska B, Kosz-Vnenchak M (2014)."General introduction into the Ebola virus biology and disease"(PDF).Folia Med Cracov.54(3): 57–65.PMID25694096.Retrieved6 August2020.
- ^Krammer F, Smith G, Fouchier R, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, García-Sastre A (28 June 2018)."Influenza".Nat Rev Dis Primers.4(1): 3.doi:10.1038/s41572-018-0002-y.PMC7097467.PMID29955068.
- ^"Vesicular stomatitis virus"(PDF).Swine Health Information Center.Center for Food Security and Public Health, College of Veterinary Medicine, Iowa State University. November 2015.Retrieved6 August2020.
- ^Kolakofsky D (April 2015)."A short biased history of RNA viruses".RNA.21(4): 667–669.doi:10.1261/rna.049916.115.PMC4371325.PMID25780183.Retrieved6 August2020.
- ^Greenwood B (12 May 2014)."The contribution of vaccination to global health: past, present and future".Philos Trans R Soc Lond B Biol Sci.369(1645): 20130433.doi:10.1098/rstb.2013.0433.PMC4024226.PMID24821919.
- ^"ICTV Taxonomy history: Negarnaviricota".International Committee on Taxonomy of Viruses (ICTV).Retrieved6 August2020.
Further reading
[edit]- Ward, C. W. (1993)."Progress towards a higher taxonomy of viruses".Research in Virology.144(6): 419–53.doi:10.1016/S0923-2516(06)80059-2.PMC7135741.PMID8140287.