Chemiosmosis
Chemiosmosisis the movement ofionsacross asemipermeable membranebound structure, down theirelectrochemical gradient.An important example is the formation ofadenosine triphosphate (ATP)by the movement ofhydrogenions (H+) across amembraneduringcellular respirationorphotosynthesis.
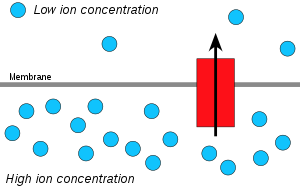
Hydrogen ions, orprotons,willdiffusefrom a region of high proton concentration to a region of lower proton concentration, and anelectrochemical concentration gradientof protons across a membrane can be harnessed to make ATP. This process is related toosmosis,the movement ofwateracross a selective membrane, which is why it is called "chemiosmosis".
ATP synthaseis theenzymethat makes ATP by chemiosmosis. It allows protons to pass through the membrane and uses thefree energydifference to convertphosphorylateadenosine diphosphate(ADP) into ATP. The ATP synthase contains two parts: CF0 (present in thylakoid membrane) and CF1 (protrudes on the outer surface of thylakoid membrane). The breakdown of the proton gradient leads to conformational change in CF1—providing enough energy in the process to convert ADP to ATP. The generation of ATP by chemiosmosis occurs inmitochondriaandchloroplasts,as well as in mostbacteriaandarchaea.For instance, inchloroplastsduring photosynthesis, an electron transport chain pumps H+ions (protons) in thestroma (fluid)through the thylakoid membrane to the thylakoid spaces. The stored energy is used tophotophosphorylateADP, making ATP, as protons move through ATP synthase.
The chemiosmotic hypothesis
[edit]Peter D. Mitchellproposed the chemiosmotic hypothesis in 1961.[1]In brief, the hypothesis was that mostadenosine triphosphate(ATP) synthesis inrespiringcells comes from theelectrochemical gradientacross the inner membranes ofmitochondriaby using theenergyofNADHandFADH2formed during the oxidative breakdown of energy-richmoleculessuch asglucose.
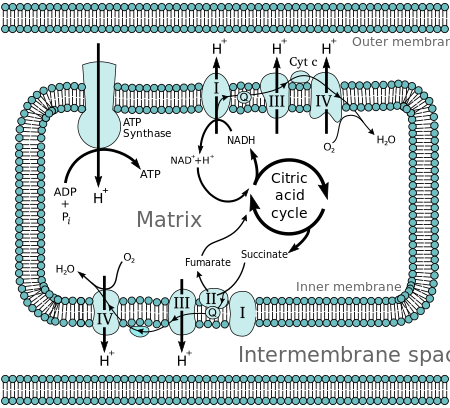
Molecules such as glucose aremetabolizedto produceacetyl CoAas a fairly energy-rich intermediate. Theoxidationofacetyl coenzyme A(acetyl-CoA) in themitochondrial matrixis coupled to thereductionof a carrier molecule such asnicotinamide adenine dinucleotide(NAD) andflavin adenine dinucleotide(FAD).[2] The carriers passelectronsto theelectron transport chain(ETC) in theinner mitochondrial membrane,which in turn pass them to other proteins in the ETC. The energy at every redox transfer step is used to pumpprotonsfrom thematrixinto the intermembrane space, storing energy in the form of a transmembraneelectrochemical gradient.The protons move back across the inner membrane through the enzymeATP synthase.The flow of protons back into the matrix of the mitochondrion viaATP synthaseprovides enough energy for ADP to combine with inorganicphosphateto form ATP.
This was a radical proposal at the time, and was not well accepted. The prevailing view was that the energy of electron transfer was stored as a stable high potential intermediate, a chemically more conservative concept. The problem with the older paradigm is that no high energy intermediate was ever found, and the evidence for proton pumping by the complexes of theelectron transfer chaingrew too great to be ignored. Eventually the weight of evidence began to favor the chemiosmotic hypothesis, and in 1978Peter D. Mitchellwas awarded theNobel Prize in Chemistry.[3]
Chemiosmotic coupling is important for ATP production inmitochondria,chloroplasts[4] and manybacteriaandarchaea.[5]
Proton-motive force
[edit]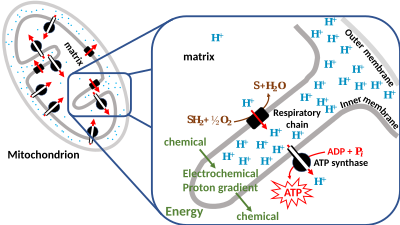
The movement of ions across the membrane depends on a combination of two factors:
- Diffusionforce caused by a concentration gradient - all particles tend to diffuse from higher concentration to lower.
- Electrostatic forcecaused byelectrical potentialgradient -cationslike protons H+tend to diffuse down the electrical potential, from the positive (P) side of the membrane to the negative (N) side.Anionsdiffuse spontaneously in the opposite direction.
These two gradients taken together can be expressed as anelectrochemical gradient.
Lipid bilayersofbiological membranes,however, are barriers for ions. This is why energy can be stored as a combination of these two gradients across the membrane. Only special membrane proteins likeion channelscan sometimes allow ions to move across the membrane (see also:Membrane transport). In the chemiosmotic hypothesis a transmembraneATP synthaseis central to convert energy of spontaneous flow of protons through them into chemical energy of ATP bonds.
Hence researchers created the termproton-motive force(PMF), derived from the electrochemical gradient mentioned earlier. It can be described as the measure of the potential energy stored (chemiosmotic potential) as a combination of proton and voltage (electrical potential) gradients across a membrane. The electrical gradient is a consequence of the charge separation across the membrane (when the protons H+move without acounterion,such aschlorideCl−).
In most cases the proton-motive force is generated by an electron transport chain which acts as a proton pump, using theGibbs free energyofredoxreactions to pump protons (hydrogen ions) out across the membrane, separating the charge across the membrane. In mitochondria, energy released by the electron transport chain is used to move protons from the mitochondrial matrix (N side) to the intermembrane space (P side). Moving the protons out of the mitochondrion creates a lower concentration of positively charged protons inside it, resulting in excess negative charge on the inside of the membrane. The electrical potential gradient is about -170 mV[6],negative inside (N). These gradients - charge difference and the proton concentration difference both create a combined electrochemical gradient across the membrane, often expressed as the proton-motive force (PMF). In mitochondria, the PMF is almost entirely made up of the electrical component but in chloroplasts the PMF is made up mostly of the pH gradient because the charge of protons H+is neutralized by the movement of Cl−and other anions. In either case, the PMF needs to be greater than about 460 mV (45 kJ/mol) for the ATP synthase to be able to make ATP.
Equations
[edit]The proton-motive force is derived from theGibbs free energy.Let N denote the inside of a cell, and P denote the outside. Then[6]
where
- is the Gibbs free energy change per unit amount ofcationstransferred from P to N;
- is thecharge numberof thecation;
- is the electric potential of N relative to P;
- andare the cation concentrations at P and N, respectively;
- is theFaraday constant;
- is thegas constant;and
- is thetemperature.
The molar Gibbs free energy changeis frequently interpreted as a molar electrochemical ion potential.
For anelectrochemical proton gradientand as a consequence:
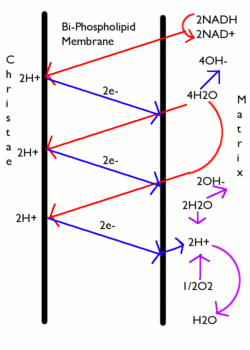
where
- .
Mitchell defined theproton-motive force(PMF) as
- .
For example,implies.Atthis equation takes the form:
.
Note that for spontaneous proton import from the P side (relatively more positive and acidic) to the N side (relatively more negative and alkaline),is negative (similar to) whereas PMF is positive (similar to redox cell potential).
It is worth noting that, as with any transmembrane transport process, the PMF is directional. The sign of the transmembrane electric potential differenceis chosen to represent the change in potential energy per unit charge flowing into the cell as above. Furthermore, due to redox-driven proton pumping by coupling sites, the proton gradient is always inside-alkaline. For both of these reasons, protons flow in spontaneously, from the P side to the N side; the available free energy is used to synthesize ATP (see below). For this reason, PMF is defined for proton import, which is spontaneous. PMF for proton export, i.e., proton pumping as catalyzed by the coupling sites, is simply the negative of PMF(import).
The spontaneity of proton import (from the P to the N side) is universal in all bioenergetic membranes.[8]This fact was not recognized before the 1990s, because the chloroplast thylakoid lumen was interpreted as an interior phase, but in fact it is topologically equivalent to the exterior of the chloroplast. Azzone et al. stressed that the inside phase (N side of the membrane) is the bacterial cytoplasm, mitochondrial matrix, or chloroplast stroma; the outside (P) side is the bacterial periplasmic space, mitochondrial intermembrane space, or chloroplast lumen. Furthermore, 3D tomography of the mitochondrial inner membrane shows its extensive invaginations to be stacked, similar to thylakoid disks; hence the mitochondrial intermembrane space is topologically quite similar to the chloroplast lumen.:[9]
The energy expressed here as Gibbs free energy, electrochemical proton gradient, or proton-motive force (PMF), is a combination of two gradients across the membrane:
- the concentration gradient (via) and
- electric potential gradient.
When a system reaches equilibrium,;nevertheless, the concentrations on either side of the membrane need not be equal. Spontaneous movement across the potential membrane is determined by both concentration and electric potential gradients.
The molar Gibbs free energyof ATP synthesis
is also called phosphorylation potential. The equilibrium concentration ratiocan be calculated by comparingand,for example in case of the mammalian mitochondrion:[9]
H+/ ATP = ΔGp/ (Δp / 10.4 kJ·mol−1/mV) = 40.2 kJ·mol−1/ (173.5 mV / 10.4 kJ·mol−1/mV) = 40.2 / 16.7 = 2.4. The actual ratio of the proton-binding c-subunit to the ATP-synthesizing beta-subunit copy numbers is 8/3 = 2.67, showing that under these conditions, the mitochondrion functions at 90% (2.4/2.67) efficiency.[9]
In fact, the thermodynamic efficiency is mostly lower in eukaryotic cells because ATP must be exported from the matrix to the cytoplasm, and ADP and phosphate must be imported from the cytoplasm. This "costs" one "extra" proton import per ATP,[6][7]hence the actual efficiency is only 65% (= 2.4/3.67).
In mitochondria
[edit]
The complete breakdown ofglucosereleasing its energy is calledcellular respiration.The last steps of this process occur in mitochondria. The reduced moleculesNADHandFADH2are generated by theKrebs cycle,glycolysis,andpyruvateprocessing. These molecules pass electrons to anelectron transport chain,which releases the energy of oxygen to create a proton gradient across the innermitochondrial membrane.ATP synthasethen uses the energy stored in this gradient to make ATP. This process is calledoxidative phosphorylationbecause it uses energy released by theoxidationof NADH and FADH2to phosphorylateADPintoATP.
In plants
[edit]Thelight reactionsofphotosynthesisgenerate ATP by the action of chemiosmosis. Thephotonsinsunlightare received by the antenna complex ofPhotosystem II,which exciteselectronsto a higherenergy level.These electrons travel down anelectron transport chain,causing protons to be actively pumped across thethylakoid membraneinto thethylakoid lumen.These protons then flow down their electrochemical potential gradient through an enzyme called ATP-synthase, creating ATP by thephosphorylationof ADP to ATP. The electrons from the initiallight reactionreachPhotosystem I,then are raised to a higher energy level by light energy and then received by an electron acceptor and reduce NADP+toNADPH.The electrons lost from Photosystem II get replaced by the oxidation of water, which is "split" into protons and oxygen by the oxygen-evolving complex (OEC, also known as WOC, or the water-oxidizing complex). To generate one molecule of diatomic oxygen, 10 photons must be absorbed by Photosystems I and II, four electrons must move through the two photosystems, and 2 NADPH are generated (later used for carbon dioxide fixation in the Calvin Cycle).
In prokaryotes
[edit]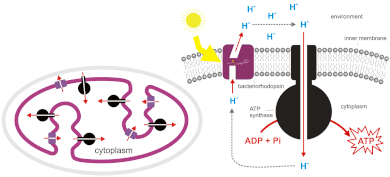
Bacteriaandarchaeaalso can use chemiosmosis to generate ATP.Cyanobacteria,green sulfur bacteria,andpurple bacteriasynthesize ATP by a process calledphotophosphorylation.[6][7]These bacteria use the energy of light to create a proton gradient using aphotosyntheticelectron transport chain.Non-photosynthetic bacteria such asE. colialso containATP synthase.In fact, mitochondria andchloroplastsare the product of endosymbiosis and trace back to incorporated prokaryotes. This process is described in theendosymbiotic theory.The origin of the mitochondrion triggered the origin of eukaryotes, and the origin of the plastid the origin of the Archaeplastida, one of the major eukaryotic supergroups.[citation needed]
Chemiosmotic phosphorylationis the third pathway that produces ATP from inorganicphosphateand an ADP molecule. This process is part of oxidative phosphorylation.
Emergence of chemiosmosis
[edit]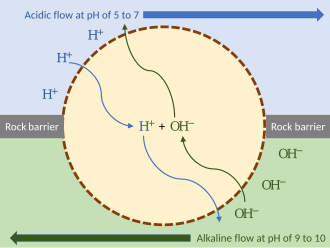
Thermal cycling model
[edit]A stepwise model for the emergence of chemiosmosis, a key element in theorigin of lifeon earth, proposes that primordial organisms used thermal cycling as an energy source (thermosynthesis), functioning essentially as a heat engine:[11]
- self-organizedconvectionin natural waters causing thermal cycling →
- added β-subunit of F1ATP Synthase
- (generated ATP by thermal cycling of subunit during suspension in convection cell: thermosynthesis) →
- added membrane and FoATP Synthase moiety
- (generated ATP by change in electrical polarization of membrane during thermal cycling: thermosynthesis) →
- added metastable, light-induced electric dipoles in membrane
- (primitive photosynthesis) →
- added quinones and membrane-spanning light-induced electric dipoles
- (today's bacterial photosynthesis, which makes use of chemiosmosis).
External proton gradient model
[edit]Deep-seahydrothermal vents,emitting hot acidic or alkaline water, would have created external proton gradients. These provided energy that primordial organisms could have exploited. To keep the flows separate, such an organism could have wedged itself in the rock of the hydrothermal vent, exposed to the hydrothermal flow on one side and the more alkaline water on the other. As long as the organism's membrane (or passiveion channelswithin it) is permeable to protons, the mechanism can function without ion pumps. Such a proto-organism could then have evolved further mechanisms such as ion pumps and ATP synthase.[10]
Meteoritic quinones
[edit]A proposed alternative source to chemiosmotic energy developing across membranous structures is if an electron acceptor, ferricyanide, is within a vesicle and the electron donor is outside, quinones transported by carbonaceous meteorites pick up electrons and protons from the donor. They would release electrons across the lipid membrane by diffusion to ferricyanide within the vesicles and release protons which produces gradients above pH 2, the process is conducive to the development of proton gradients.[12][13]
See also
[edit]- Cellular respiration
- Citric acid cycle
- Electrochemical gradient
- Glycolysis
- Oxidative phosphorylation
References
[edit]- ^Mitchell P (July 1961). "Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism".Nature.191(4784): 144–148.Bibcode:1961Natur.191..144M.doi:10.1038/191144a0.PMID13771349.S2CID1784050.
- ^Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002)."Proton Gradients Produce Most of the Cell's ATP".Molecular Biology of the Cell.Garland.ISBN0-8153-4072-9.
- ^TheNobel Prizein Chemistry 1978.
- ^Cooper GM (2000)."Figure 10.22: Electron transport and ATP synthesis during photosynthesis".The Cell: A Molecular Approach(2nd ed.). Sinauer Associates, Inc.ISBN0-87893-119-8.
- ^Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002)."Figure 14-32: The importance of H+-driven transport in bacteria ".Molecular Biology of the Cell.Garland.ISBN0-8153-4072-9.
- ^abcdefgNicholls D. G.;Ferguson S. J. (1992).Bioenergetics 2(2nd ed.). San Diego: Academic Press.ISBN9780125181242.
- ^abcdStryer L (1995).Biochemistry(fourth ed.). New York - Basingstoke: W. H. Freeman and Company.ISBN978-0716720096.
- ^Azzone G, Benz R, Bertl A, Colombini M, Crofts A, Dilley R, Dimroth P, Dutton PL, Felle H, Harold F, Junge W (1993). "Transmembrane Measurements Across Bioenergetic Membranes".Biochimica et Biophysica Acta (BBA) - Bioenergetics.1183(1): 1–3.doi:10.1016/0005-2728(93)90002-W.
- ^abcSilverstein TP (June 2014). "An exploration of how the thermodynamic efficiency of bioenergetic membrane systems varies with c-subunit stoichiometry of F₁F₀ ATP synthases".Journal of Bioenergetics and Biomembranes.46(3): 229–241.doi:10.1007/s10863-014-9547-y.PMID24706236.S2CID1840860.
- ^abLane N(2015).The Vital Question: Why Is Life The Way It Is?.Profile Books. pp. 129–140.ISBN978-1781250365.
- ^Muller AW (2012). "Life Explained by Heat Engines". In Seckbach J (ed.).Genesis — in the Beginning.Cellular Origin, Life in Extreme Habitats and Astrobiology. Vol. 22. Springer. pp. 321–344.doi:10.1007/978-94-007-2941-4_19.ISBN978-94-007-2940-7.
- ^Damer B, Deamer D (April 2020)."The Hot Spring Hypothesis for an Origin of Life".Astrobiology.20(4): 429–452.Bibcode:2020AsBio..20..429D.doi:10.1089/ast.2019.2045.PMC7133448.PMID31841362.
- ^Milshteyn D, Cooper G, Deamer D (August 2019)."Chemiosmotic energy for primitive cellular life: Proton gradients are generated across lipid membranes by redox reactions coupled to meteoritic quinones".Scientific Reports.9(1): 12447.Bibcode:2019NatSR...912447M.doi:10.1038/s41598-019-48328-5.PMC6713726.PMID31462644.
Further reading
[edit]- Biochemistry textbook reference, from theNCBI bookshelf–Jeremy M. Berg; John L. Tymoczko; Lubert Stryer (eds.). "18.4. A Proton Gradient Powers the Synthesis of ATP".Biochemistry(5th ed.). W. H. Freeman.
- A set of experiments aiming to test some tenets of the chemiosmotic theory–Ogawa S, Lee TM (August 1984)."The relation between the internal phosphorylation potential and the proton motive force in mitochondria during ATP synthesis and hydrolysis".The Journal of Biological Chemistry.259(16): 10004–10011.doi:10.1016/S0021-9258(18)90918-X.PMID6469951.

![{\displaystyle \Delta \!G=zF\Delta \!\psi +RT\ln {\frac {[\mathrm {X} ^{z+}]_{\text{N}}}{[\mathrm {X} ^{z+}]_{\text{P}}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/e49984fb465bfe70fdf147d5c94b4691fde30b93)




![{\displaystyle [\mathrm {X} ^{z+}]_{\text{P}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/b04667620cf542631c6a2a692aefd89310230ddd)
![{\displaystyle [\mathrm {X} ^{z+}]_{\text{N}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/f4da81b44fd243b0610ca90f2a571ffe2891ad71)





![{\displaystyle \Delta \!\mu _{\mathrm {H} ^{+}}=F\Delta \!\psi +RT\ln {\frac {[\mathrm {H} ^{+}]_{\text{N}}}{[\mathrm {H} ^{+}]_{\text{P}}}}=F\Delta \!\psi -(\ln 10)RT\Delta \mathrm {pH} }](https://wikimedia.org/api/rest_v1/media/math/render/svg/9401e1568170355be3a960a583f16f2d93c5a842)













![{\displaystyle [\mathrm {H} ^{+}]/[\mathrm {ATP} ]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/0e253bb9b194bd7dd88012ff9e7acd912dbf6931)
