DNA-binding protein



DNA-binding proteinsareproteinsthat haveDNA-binding domainsand thus have a specific or general affinity forsingle- or double-stranded DNA.[3][4][5]Sequence-specific DNA-binding proteins generally interact with themajor grooveofB-DNA,because it exposes morefunctional groupsthat identify abase pair.[6][7]
Examples[edit]
DNA-bindingproteinsincludetranscription factorswhichmodulatethe process of transcription, variouspolymerases,nucleaseswhich cleave DNA molecules, andhistoneswhich are involved inchromosomepackaging and transcription in thecell nucleus.DNA-binding proteins can incorporate such domains as thezinc finger,thehelix-turn-helix,and theleucine zipper(among many others) that facilitate binding to nucleic acid. There are also more unusual examples such astranscription activator like effectors.
Non-specific DNA-protein interactions[edit]
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure calledchromatin.Ineukaryotes,this structure involves DNA binding to a complex of small basic proteins calledhistones.Inprokaryotes,multiple types of proteins are involved.[8][9]The histones form a disk-shaped complex called anucleosome,which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones makingionic bondsto the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.[10]Chemicalmodifications of these basicamino acidresidues includemethylation,phosphorylationandacetylation.[11]These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible totranscription factorsand changing the rate of transcription.[12]Other non-specific DNA-binding proteins in chromatin include the high-mobility group (HMG) proteins, which bind to bent or distorted DNA.[13]Biophysical studies show that these architectural HMG proteins bind, bend and loop DNA to perform its biological functions.[14][15]These proteins are important in bending arrays of nucleosomes and arranging them into the larger structures that form chromosomes.[16]Recently FK506 binding protein 25 (FBP25) was also shown to non-specifically bind to DNA which helps in DNA repair.[17]
Proteins that specifically bind single-stranded DNA[edit]
A distinct group of DNA-binding proteins are the DNA-binding proteins that specifically bind single-stranded DNA. In humans,replication protein Ais the best-understood member of this family and is used in processes where the double helix is separated, including DNA replication, recombination and DNA repair.[18]These binding proteins seem to stabilize single-stranded DNA and protect it from formingstem-loopsor being degraded bynucleases.
Binding to specific DNA sequences[edit]
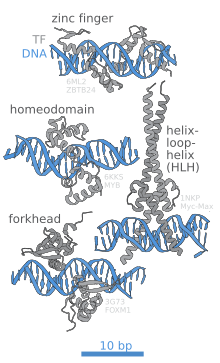
In contrast, other proteins have evolved to bind to specific DNA sequences. The most intensively studied of these are the varioustranscription factors,which are proteins that regulate transcription. Each transcription factor binds to one specific set of DNA sequences and activates or inhibits the transcription of genes that have these sequences near their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[19]Alternatively, transcription factors can bindenzymesthat modify the histones at the promoter. This alters the accessibility of the DNA template to the polymerase.[20]
These DNA targets can occur throughout an organism's genome. Thus, changes in the activity of one type of transcription factor can affect thousands of genes.[21]Thus, these proteins are often the targets of thesignal transductionprocesses that control responses to environmental changes orcellular differentiationand development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them toreadthe DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most accessible.[22]Mathematical descriptions of protein-DNA binding taking into account sequence-specificity, and competitive and cooperative binding of proteins of different types are usually performed with the help of thelattice models.[23]Computational methods to identify the DNA binding sequence specificity have been proposed to make a good use of the abundant sequence data in the post-genomic era.[24]
Protein–DNA interactions[edit]
Protein–DNA interactions occur when aproteinbinds a molecule ofDNA,often to regulate thebiological functionof DNA, usually theexpressionof agene.Among the proteins that bind to DNA aretranscription factorsthat activate or repress gene expression by binding to DNA motifs andhistonesthat form part of the structure of DNA and bind to it less specifically. Also proteins thatrepair DNAsuch asuracil-DNA glycosylaseinteract closely with it.
In general, proteins bind to DNA in themajor groove;however, there are exceptions.[25]Protein–DNA interaction are of mainly two types, either specific interaction, or non-specific interaction. Recent single-molecule experiments showed that DNA binding proteins undergo of rapid rebinding in order to bind in correct orientation for recognizing the target site.[26]
Design[edit]
Designing DNA-binding proteins that have a specified DNA-binding site has been an important goal for biotechnology.Zinc fingerproteins have been designed to bind to specific DNA sequences and this is the basis ofzinc finger nucleases.Recentlytranscription activator-like effector nucleases(TALENs) have been created which are based on naturalproteinssecreted byXanthomonasbacteria via theirtype III secretion systemwhen they infect variousplantspecies.[27]
Detection methods[edit]
There are manyin vitroandin vivotechniques which are useful in detecting DNA-Protein Interactions. The following lists some methods currently in use:[28]Electrophoretic mobility shift assay(EMSA) is a widespread qualitative technique to study protein–DNA interactions of known DNA binding proteins.[29][30]DNA-Protein-Interaction - Enzyme-Linked ImmunoSorbant Assay (DPI-ELISA)allows the qualitative and quantitative analysis of DNA-binding preferences of known proteinsin vitro.[31][32]This technique allows the analysis of protein complexes that bind to DNA (DPI-Recruitment-ELISA) or is suited for automated screening of several nucleotide probes due to its standard ELISA plate formate[33][34].DNase footprinting assaycan be used to identify the specific sites of binding of a protein to DNA at basepair resolution.[35]Chromatin immunoprecipitationis used to identify thein vivoDNA target regions of a known transcription factor. This technique when combined with high throughput sequencing is known asChIP-Seqand when combined withmicroarraysit is known asChIP-chip.Yeast one-hybrid System(Y1H) is used to identify which protein binds to a particular DNA fragment.Bacterial one-hybrid system(B1H) is used to identify which protein binds to a particular DNA fragment. Structure determination usingX-ray crystallographyhas been used to give a highly detailed atomic view of protein–DNA interactions. Besides these methods, other techniques such as SELEX, PBM (protein binding microarrays), DNA microarray screens, DamID, FAIRE or more recently DAP-seq are used in the laboratory to investigate DNA-protein interactionin vivoandin vitro.
Manipulating the interactions[edit]
The protein–DNA interactions can be modulated using stimuli like ionic strength of the buffer, macromolecular crowding,[26]temperature, pH and electric field. This can lead to reversible dissociation/association of the protein–DNA complex.[36][37]
See also[edit]
- bZIP domain
- ChIP-exo
- Comparison of nucleic acid simulation software
- DNA-binding domain
- Helix-loop-helix
- Helix-turn-helix
- HMG-box
- Leucine zipper
- Lexitropsin(a semi-synthetic DNA-binding ligand)
- Deoxyribonucleoprotein
- Protein–DNA interaction site prediction software
- RNA-binding protein
- Single-strand binding protein
- Zinc finger
References[edit]
- ^Created fromPDB 1LMB
- ^Created fromPDB 1RVA
- ^Travers, A. A. (1993).DNA-protein interactions.London: Springer.ISBN978-0-412-25990-6.
- ^Pabo CO, Sauer RT (1984). "Protein-DNA recognition".Annu. Rev. Biochem.53(1): 293–321.doi:10.1146/annurev.bi.53.070184.001453.PMID6236744.
- ^Dickerson R.E. (1983). "The DNA helix and how it is read".Sci Am.249(6): 94–111.Bibcode:1983SciAm.249f..94D.doi:10.1038/scientificamerican1283-94.
- ^Zimmer C, Wähnert U (1986)."Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material".Prog. Biophys. Mol. Biol.47(1): 31–112.doi:10.1016/0079-6107(86)90005-2.PMID2422697.
- ^Dervan PB (April 1986). "Design of sequence-specific DNA-binding molecules".Science.232(4749): 464–71.Bibcode:1986Sci...232..464D.doi:10.1126/science.2421408.PMID2421408.
- ^Sandman K, Pereira S, Reeve J (1998)."Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome".Cell Mol Life Sci.54(12): 1350–64.doi:10.1007/s000180050259.PMC11147202.PMID9893710.S2CID21101836.
- ^Dame RT (2005). "The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin".Mol. Microbiol.56(4): 858–70.doi:10.1111/j.1365-2958.2005.04598.x.PMID15853876.
- ^Luger K, Mäder A, Richmond R, Sargent D, Richmond T (1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution".Nature.389(6648): 251–60.Bibcode:1997Natur.389..251L.doi:10.1038/38444.PMID9305837.S2CID4328827.
- ^Jenuwein T, Allis C (2001). "Translating the histone code".Science.293(5532): 1074–80.CiteSeerX10.1.1.453.900.doi:10.1126/science.1063127.PMID11498575.S2CID1883924.
- ^Ito T (2003). "Nucleosome Assembly and Remodeling".Protein Complexes that Modify Chromatin.Current Topics in Microbiology and Immunology. Vol. 274. pp. 1–22.doi:10.1007/978-3-642-55747-7_1.ISBN978-3-642-62909-9.PMID12596902.
{{cite book}}:|journal=ignored (help) - ^Thomas J (2001). "HMG1 and 2: architectural DNA-binding proteins".Biochem Soc Trans.29(Pt 4): 395–401.doi:10.1042/BST0290395.PMID11497996.
- ^Murugesapillai, Divakaran; McCauley, Micah J.; Huo, Ran; Nelson Holte, Molly H.; Stepanyants, Armen; Maher, L. James; Israeloff, Nathan E.; Williams, Mark C. (2014)."DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin".Nucleic Acids Research.42(14): 8996–9004.doi:10.1093/nar/gku635.PMC4132745.PMID25063301.
- ^Murugesapillai, Divakaran; McCauley, Micah J.; Maher, L. James; Williams, Mark C. (2017)."Single-molecule studies of high-mobility group B architectural DNA bending proteins".Biophysical Reviews.9(1): 17–40.doi:10.1007/s12551-016-0236-4.PMC5331113.PMID28303166.
- ^Grosschedl R, Giese K, Pagel J (1994). "HMG domain proteins: architectural elements in the assembly of nucleoprotein structures".Trends Genet.10(3): 94–100.doi:10.1016/0168-9525(94)90232-1.PMID8178371.
- ^Prakash, Ajit; Shin, Joon; Rajan, Sreekanth; Yoon, Ho Sup (2016-04-07)."Structural basis of nucleic acid recognition by FK506-binding protein 25 (FKBP25), a nuclear immunophilin".Nucleic Acids Research.44(6): 2909–2925.doi:10.1093/nar/gkw001.ISSN0305-1048.PMC4824100.PMID26762975.
- ^Iftode C, Daniely Y, Borowiec J (1999). "Replication protein A (RPA): the eukaryotic SSB".Crit Rev Biochem Mol Biol.34(3): 141–80.doi:10.1080/10409239991209255.PMID10473346.
- ^Myers L, Kornberg R (2000). "Mediator of transcriptional regulation".Annu Rev Biochem.69(1): 729–49.doi:10.1146/annurev.biochem.69.1.729.PMID10966474.
- ^Spiegelman B, Heinrich R (2004)."Biological control throughs regulated transcriptional coactivators".Cell.119(2): 157–67.doi:10.1016/j.cell.2004.09.037.PMID15479634.S2CID14668705.
- ^Li Z, Van Calcar S, Qu C, Cavenee W, Zhang M, Ren B (2003)."A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells".Proc Natl Acad Sci USA.100(14): 8164–9.Bibcode:2003PNAS..100.8164L.doi:10.1073/pnas.1332764100.PMC166200.PMID12808131.
- ^Pabo C, Sauer R (1984). "Protein-DNA recognition".Annu Rev Biochem.53(1): 293–321.doi:10.1146/annurev.bi.53.070184.001453.PMID6236744.
- ^Teif V.B.; Rippe K. (2010). "Statistical-mechanical lattice models for protein-DNA binding in chromatin".Journal of Physics: Condensed Matter.22(41): 414105.arXiv:1004.5514.Bibcode:2010JPCM...22O4105T.doi:10.1088/0953-8984/22/41/414105.PMID21386588.S2CID103345.
- ^Wong KC, Chan TM, Peng C, Li Y, Zhang Z (2013)."DNA Motif Elucidation using belief propagation".Nucleic Acids Research.41(16): e153.doi:10.1093/nar/gkt574.PMC3763557.PMID23814189.
- ^Bewley CA, Gronenborn AM, Clore GM (1998)."Minor groove-binding architectural proteins: structure, function, and DNA recognition".Annu Rev Biophys Biomol Struct.27:105–31.doi:10.1146/annurev.biophys.27.1.105.PMC4781445.PMID9646864.
- ^abGanji, Mahipal; Docter, Margreet; Le Grice, Stuart F. J.; Abbondanzieri, Elio A. (2016-09-30)."DNA binding proteins explore multiple local configurations during docking via rapid rebinding".Nucleic Acids Research.44(17): 8376–8384.doi:10.1093/nar/gkw666.ISSN0305-1048.PMC5041478.PMID27471033.
- ^Clark KJ, Voytas DF, Ekker SC (September 2011)."A TALE of two nucleases: gene targeting for the masses?".Zebrafish.8(3): 147–9.doi:10.1089/zeb.2011.9993.PMC3174730.PMID21929364.
- ^Cai YH, Huang H (July 2012). "Advances in the study of protein–DNA interaction".Amino Acids.43(3): 1141–6.doi:10.1007/s00726-012-1377-9.PMID22842750.S2CID310256.
- ^Fried M, Crothers DM (1981)."Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis".Nucleic Acids Res.9(23): 6505–6525.doi:10.1093/nar/9.23.6505.PMC327619.PMID6275366.
- ^Garner MM, Revzin A (1981)."A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system".Nucleic Acids Res.9(13): 3047–3060.doi:10.1093/nar/9.13.3047.PMC327330.PMID6269071.
- ^Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D (2010)."DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro".Plant Methods.25(6): 25.doi:10.1186/1746-4811-6-25.PMC3003642.PMID21108821.
- ^Fischer SM, Böser A, Hirsch JP, Wanke D (2016). "Quantitative Analysis of Protein–DNA Interaction by qDPI-ELISA".Plant Synthetic Promoters.Methods Mol. Biol. Vol. 1482. pp. 49–66.doi:10.1007/978-1-4939-6396-6_4.ISBN978-1-4939-6394-2.PMID27557760.
- ^Hecker A, Brand LH, Peter S, Simoncello N, Kilian J, Harter K, Gaudin V, Wanke D (2015)."The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs".Plant Physiol.163(3): 1013–1024.doi:10.1104/pp.15.00409.PMC4741334.PMID26025051.
- ^Brand LH, Henneges C, Schüssler A, Kolukisaoglu HÜ, Koch G, Wallmeroth N, Hecker A, Thurow K, Zell A, Harter K, Wanke D (2013)."Screening for protein-DNA interactions by automatable DNA-protein interaction ELISA".PLOS ONE.8(10): e75177.doi:10.1371/journal.pone.0075177.PMC3795721.PMID24146751.
- ^Galas DJ, Schmitz A (1978)."DNAse footprinting: a simple method for the detection of protein-DNA binding specificity".Nucleic Acids Res.5(9): 3157–3170.doi:10.1093/nar/5.9.3157.PMC342238.PMID212715.
- ^Hianik T, Wang J (2009). "Electrochemical Aptasensors – Recent Achievements and Perspectives".Electroanalysis.21(11): 1223–1235.doi:10.1002/elan.200904566.
- ^Gosai A, et al. (2016)."Electrical Stimulus Controlled Binding/Unbinding of Human Thrombin-Aptamer Complex".Sci. Rep.6:37449.Bibcode:2016NatSR...637449G.doi:10.1038/srep37449.PMC5118750.PMID27874042.
External links[edit]
- Protein-DNA binding: data, tools & models (annotated list, constantly updated)
- Abalonetool for modeling DNA-ligand interactions.
- DBD database of predicted transcription factorsUses a curated set of DNA-binding domains to predict transcription factors in all completely sequenced genomes
- DNA-Binding+Proteinsat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
