DNA methyltransferase
| N-6 DNA Methylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of type i restriction enzyme ecoki m protein (ec 2.1.1.72) (m.ecoki) | |||||||||
| Identifiers | |||||||||
| Symbol | N6_Mtase | ||||||||
| Pfam | PF02384 | ||||||||
| Pfamclan | CL0063 | ||||||||
| InterPro | IPR003356 | ||||||||
| PROSITE | PDOC00087 | ||||||||
| |||||||||
| HsdM N-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HsdM_N | ||||||||
| Pfam | PF12161 | ||||||||
| |||||||||
| C-5 cytosine-specific DNA methylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of human dnmt2, an enigmatic dna methyltransferase homologue | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_methylase | ||||||||
| Pfam | PF00145 | ||||||||
| Pfamclan | CL0063 | ||||||||
| InterPro | IPR001525 | ||||||||
| PROSITE | PDOC00089 | ||||||||
| SCOP2 | 1hmy/SCOPe/SUPFAM | ||||||||
| CDD | cd00315 | ||||||||
| |||||||||
| DNA methylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of methyltransferase mboiia (moraxella bovis) | |||||||||
| Identifiers | |||||||||
| Symbol | N6_N4_Mtase | ||||||||
| Pfam | PF01555 | ||||||||
| Pfamclan | CL0063 | ||||||||
| InterPro | IPR002941 | ||||||||
| PROSITE | PDOC00088 | ||||||||
| SCOP2 | 1boo/SCOPe/SUPFAM | ||||||||
| |||||||||
Inbiochemistry,theDNA methyltransferase(DNA MTase,DNMT) family ofenzymescatalyzethe transfer of amethyl grouptoDNA.DNA methylationserves a wide variety of biological functions. All the known DNA methyltransferases useS-adenosyl methionine(SAM) as the methyl donor.
Classification[edit]
Substrate[edit]
MTases can be divided into three different groups on the basis of the chemical reactions they catalyze:
- m6A - those that generateN6-methyladenineEC2.1.1.72
- m4C - those that generateN4-methylcytosineEC2.1.1.113
- m5C - those that generateC5-methylcytosineEC2.1.1.37
m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes[1]). m5C methyltransferases are found in some lower eukaryotes, in most higher plants, and in animals beginning with theechinoderms.
The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) areenzymesthat specifically methylate the amino group at the C-6 position ofadeninesin DNA. They are found in the three existing types ofbacterialrestriction-modification systems(in type I system the A-Mtase is theproductof the hsdM gene, and in type III it is theproductof the mod gene). Theseenzymesare responsible for themethylationof specific DNAsequencesin order to prevent the host from digesting its owngenomevia itsrestriction enzymes.These methylases have the samesequencespecificity as their corresponding restriction enzymes. These enzymes contain aconservedmotifAsp/Asn-Pro-Pro-Tyr/Phein their N-terminal section, thisconservedregion could be involved insubstratebindingor in thecatalyticactivity.[2][3][4][5]Thestructureof N6-MTase TaqI (M.TaqI) has been resolved to 2.4A.Themoleculefoldsinto 2 domains, an N-terminal catalytic domain, which contains thecatalyticandcofactorbinding sites, and comprises a central 9-stranded beta-sheet, surrounded by 5 helices; and a C-terminal DNA recognition domain, which is formed by 4 smallbeta-sheetsand 8alpha-helices.The N- and C-terminaldomainsform a cleft that accommodates the DNAsubstrate.[6]A classification of N-MTases has been proposed, based onconservedmotif(CM) arrangements.[5]According to this classification, N6-MTases that have a DPPY motif (CM II) occurring after the FxGxG motif (CM I) are designated D12 class N6-adenine MTases. The type I restriction and modification system is composed of threepolypeptidesR, M and S. The M (hsdM) and Ssubunitstogether form amethyltransferasethatmethylatestwoadenineresiduesincomplementarystrands of a bipartite DNArecognition sequence.In the presence of the R subunit, thecomplexcan also act as anendonuclease,binding to the same targetsequencebut cutting the DNA some distance from this site. Whether the DNA is cut or modified depends on the methylation state of the targetsequence.When the target site is unmodified, the DNA is cut. When the target site is hemimethylated, the complex acts as a maintenance methyltransferase, modifying the DNA so that both strands becomemethylated.hsdM contains analpha-helicaldomainat theN-terminus,the HsdM N-terminal domain.[7]
Among the m6A methyltransferases (N-6 adenine-specific DNA methylase) there is a group of orphan MTases that do not participate in the bacterial restriction/methylation system.[8]These enzymes have a regulatory role in gene expression and cell cycle regulation.EcoDamfromE. coli[9]and CcrM fromCaulobacter crescentus[10]are well characterized members of these family. More recently, CamA fromClostridioides difficile,was shown to play key functional roles insporulation,biofilmformations and host-adaptation.[11]
m4C methyltransferases (N-4 cytosine-specific DNA methylases) areenzymesthat specifically methylate the amino group at the C-4 position ofcytosinesin DNA.[5]Suchenzymesare found as components of type II restriction-modification systems inprokaryotes.Such enzymes recognise a specificsequencein DNA and methylate acytosinein thatsequence.By this action they protect DNA fromcleavageby type II restriction enzymes that recognise the samesequence[citation needed]
m5C methyltransferases (C-5 cytosine-specific DNA methylase) (C5 Mtase) are enzymes that specifically methylate the C-5carbonofcytosinesin DNA to produceC5-methylcytosine.[12][13][14]Inmammaliancells, cytosine-specificmethyltransferasesmethylate certainCpGsequences, which are believed to modulategene expressionandcell differentiation.Inbacteria,theseenzymesare a component of restriction-modification systems and serve as valuable tools for the manipulation of DNA.[13][15]Thestructureof HhaI methyltransferase (M.HhaI) has been resolved to 2.5A:the molecule folds into 2domains- a largercatalyticdomain containing catalytic andcofactorbinding sites, and a smaller DNA recognition domain.[16]
Highly conserved DNA methyltransferases of the m4C, m5C, and m6A types have been reported,[17]which appear as promising targets for the development of novel epigenetic inhibitors to fight bacterial virulence, antibiotic resistance, among other biomedical applications.
De novo vs. maintenance[edit]
De novomethyltransferases recognize something in the DNA that allows them to newly methylate cytosines. These are expressed mainly in early embryo development and they set up the pattern of methylation.De novomethyltransferases are also active when a signal-responsive cell, such as aneuron,needs to alter protein expression.[18]As an example, whenfear conditioningcreates a newmemoryin a rat, 9.17% of the genes in the rathippocampusneuron genome are differentially methylated.[19]
Maintenance methyltransferasesadd methylation to DNA when one strand is already methylated. These work throughout the life of the organism to maintain the methylation pattern that had been established by the de novo methyltransferases.[citation needed]
Mammalian[edit]
At least four differently active DNA methyltransferases have been identified in mammals. They are namedDNMT1,[20]two isoforms transcribed from theDNMT3agene: DNMT3a1 and DNMT3a2,[21]andDNMT3b.[22]Recently, another enzyme DNMT3c has been discovered specifically expressed in the male germline in the mouse.[23]

Manzo et al.[24]observed differences in genomic binding of DNMT3a1, DNMT3a2 and DNMT3b. They found 3,970 regions exclusively enriched for DNMT3a1, 3,838 exclusively enriched for DNMT3a2 and 3,432 exclusively enriched for DNMT3b.
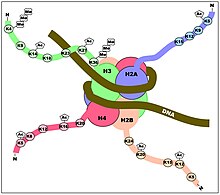
The DNMT enzymes are not only regulated in their methylating locations on the genome by where they bind to DNA,[24]but they are also regulated by thepost-translational modificationson thehistoneproteins of thenucleosomesaround which the genomic DNA is wrapped (see Figures). Rose and Klose[25]reviewed the relationship between DNA methylation andhistonelysinemethylation. For example, they indicated that H3K4me3 appears to block DNA methylation while H3K9me3 plays a role in promoting DNA methylation.
DNMT3L[26]is a protein closely related to DNMT3a and DNMT3b in structure and critical for DNA methylation, but appears to be inactive on its own.
DNMT1[edit]
DNMT1is the most abundant DNA methyltransferase in mammalian cells, and considered to be the key maintenance methyltransferase inmammals.It predominantlymethylateshemimethylatedCpGdi-nucleotides in the mammalian genome. The recognition motif for the human enzyme involves only three of the bases in the CpG dinucleotide pair: a C on one strand and CpG on the other. This relaxed substrate specificity requirement allows it to methylate unusual structures like DNA slippage intermediates at de novo rates that equal its maintenance rate.[27]Like other DNA cytosine-5 methyltransferases the human enzyme recognizes flipped out cytosines in double stranded DNA and operates by the nucleophilic attack mechanism.[28]In human cancer cells DNMT1 is responsible for bothde novoand maintenance methylation of tumor suppressor genes.[29][30]Theenzymeis about 1,620amino acidslong. The first 1,100 amino acids constitute the regulatory domain of the enzyme, and the remaining residues constitute the catalytic domain. These are joined byGly-Lysrepeats. Both domains are required for the catalytic function of DNMT1.[citation needed]
DNMT1 has severalisoforms,the somatic DNMT1, a splice variant (DNMT1b) and anoocyte-specific isoform (DNMT1o). DNMT1o is synthesized and stored in thecytoplasmof the oocyte and translocated to thecell nucleusduring earlyembryonicdevelopment, while the somatic DNMT1 is always found in the nucleus ofsomatictissue.[citation needed]
DNMT1 null mutantembryonic stem cellswere viable and contained a small percentage of methylated DNA and methyltransferase activity. Mouse embryos homozygous for a deletion in Dnmt1 die at 10–11 days gestation.[31]
TRDMT1[edit]
Although this enzyme has strong sequence similarities with 5-methylcytosine methyltransferases of both prokaryotes and eukaryotes, in 2006, the enzyme was shown to methylate position 38 inaspartic acidtransfer RNA and does not methylate DNA.[32]The name for this methyltransferase has been changed from DNMT2 to TRDMT1 (tRNA aspartic acid methyltransferase 1) to better reflect its biological function.[33]TRDMT1 is the first RNA cytosine methyltransferase to be identified in human cells.
DNMT3[edit]
DNMT3is a family ofDNAmethyltransferases that could methylate hemimethylated and unmethylatedCpGat the same rate. The architecture of DNMT3 enzymes is similar to that of DNMT1, with a regulatory region attached to a catalytic domain. There are at least five members of the DNMT3 family: DNMT3a1, DNMT3a2, 3b, 3c and 3L.[citation needed]
DNMT3a1, DNMT3a2 and DNMT3b can mediate methylation ofCpG sitesin gene promoters, resulting ingene repression.These DNA methyltransferases can also methylate CpG sites within the coding regions of genes, where such methylation can increase gene transcription.[34]Work with DNMT3a1 showed it preferentially localized to CpG islands bivalently marked by H3K4me3 (a transcription promoting mark) and H3K27me3 (a transcription repressive mark), coinciding with thepromotersof manytranscription factors.Work with DNMT3a2, inneurons,found that the DNA methylation changes caused by DNMT3a2 predominantly occur in intergenic and intronic regions. These intergenic and intronic DNA methylations were thought to likely regulateenhanceractivity,alternative splicingor the expression ofnon-coding RNAs.[35]
DNMT3a1 can co-localize withheterochromatinprotein (HP1) and methyl-CpG-binding protein (MeCBP), among a number of other factors.[36]They can also interact with DNMT1, which might be a co-operative event during DNA methylation. DNMT3a prefersCpGmethylationto CpA, CpT, and CpC methylation, though there appears to be some sequence preference of methylation for DNMT3a and DNMT3b. DNMT3a methylatesCpGsites at a rate much slower than DNMT1, but greater than DNMT3b.
The expression of DNMT3a2 differs from DNMT3a1 and DNMT3b because DNMT3a2 expression occurs in the pattern of animmediate early gene.DNMT3a2 is induced to express in neurons, for instance, by new neuronal activity.[37][35]This may be of importance in establishing long-termmemory.[38]In a rat, high levels of new DNA methylations inneuronsof thehippocampusoccur after a memorable event is imposed on a rat, such as contextualfear conditioning.[19]Bayraktar and Kreutz[39]found that DNMT inhibitors, applied in the brain, prevented long-term memories from forming.
DNMT3L contains DNA methyltransferasemotifsand is required for establishing maternalgenomic imprints,despite beingcatalyticallyinactive. DNMT3L is expressed duringgametogenesiswhengenomic imprintingtakes place. The loss of DNMT3L leads to bi-allelicexpression of genes normally not expressed by the maternal allele. DNMT3L interacts with DNMT3a and DNMT3b and co-localized in the nucleus. Though DNMT3L appears incapable ofmethylation,it may participate intranscriptionalrepression.
Clinical significance[edit]
DNMT inhibitors[edit]
Because of theepigenetic effectsof the DNMT family, someDNMT inhibitorsare under investigation for treatment of some cancers:[40]
- Vidaza(azacitidine) in phase III trials formyelodysplastic syndromesandAML
- Dacogen(decitabine) in phase III trials for AML andCML.EUapproved in 2012 for AML.[41]
- guadecitabine, an experimental drug under development by Astex Pharmaceuticals and Otsuka Pharmaceutical. It failed to meet primary endpoints in a 2018 Phase III AML trial.
See also[edit]
References[edit]
- ^Iyer LM, Zhang D, Aravind L (January 2016)."Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification".BioEssays.38(1): 27–40.doi:10.1002/bies.201500104.PMC4738411.PMID26660621.
- ^Loenen WA, Daniel AS, Braymer HD, Murray NE (November 1987). "Organization and sequence of the hsd genes of Escherichia coli K-12".Journal of Molecular Biology.198(2): 159–70.doi:10.1016/0022-2836(87)90303-2.PMID3323532.
- ^Narva KE, Van Etten JL, Slatko BE, Benner JS (December 1988). "The amino acid sequence of the eukaryotic DNA [N6-adenine]methyltransferase, M.CviBIII, has regions of similarity with the prokaryotic isoschizomer M.TaqI and other DNA [N6-adenine] methyltransferases".Gene.74(1): 253–9.doi:10.1016/0378-1119(88)90298-3.PMID3248728.
- ^Lauster R (March 1989). "Evolution of type II DNA methyltransferases. A gene duplication model".Journal of Molecular Biology.206(2): 313–21.doi:10.1016/0022-2836(89)90481-6.PMID2541254.
- ^abcTiminskas A, Butkus V, Janulaitis A (May 1995). "Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases".Gene.157(1–2): 3–11.doi:10.1016/0378-1119(94)00783-O.PMID7607512.
- ^Labahn J, Granzin J, Schluckebier G, Robinson DP, Jack WE, Schildkraut I, Saenger W (November 1994)."Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine".Proceedings of the National Academy of Sciences of the United States of America.91(23): 10957–61.doi:10.1073/pnas.91.23.10957.PMC45145.PMID7971991.
- ^Kelleher JE, Daniel AS, Murray NE (September 1991). "Mutations that confer de novo activity upon a maintenance methyltransferase".Journal of Molecular Biology.221(2): 431–40.doi:10.1016/0022-2836(91)80064-2.PMID1833555.
- ^Adhikari S, Curtis PD (September 2016)."DNA methyltransferases and epigenetic regulation in bacteria".FEMS Microbiology Reviews.40(5): 575–91.doi:10.1093/femsre/fuw023.PMID27476077.
- ^Chahar S, Elsawy H, Ragozin S, Jeltsch A (January 2010). "Changing the DNA recognition specificity of the EcoDam DNA-(adenine-N6)-methyltransferase by directed evolution".Journal of Molecular Biology.395(1): 79–88.doi:10.1016/j.jmb.2009.09.027.PMID19766657.
- ^Maier JA, Albu RF, Jurkowski TP, Jeltsch A (December 2015). "Investigation of the C-terminal domain of the bacterial DNA-(adenine N6)-methyltransferase CcrM".Biochimie.119:60–7.doi:10.1016/j.biochi.2015.10.011.PMID26475175.
- ^Oliveira PH, Ribis JW, Garrett EM, Trzilova D, Kim A, Sekulovic O, et al. (January 2020)."Epigenomic characterization of Clostridioides difficile finds a conserved DNA methyltransferase that mediates sporulation and pathogenesis".Nature Microbiology.5(1): 166–180.doi:10.1038/s41564-019-0613-4.PMC6925328.PMID31768029.
- ^Pósfai J, Bhagwat AS, Roberts RJ (December 1988). "Sequence motifs specific for cytosine methyltransferases".Gene.74(1): 261–5.doi:10.1016/0378-1119(88)90299-5.PMID3248729.
- ^abKumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG (January 1994)."The DNA (cytosine-5) methyltransferases".Nucleic Acids Research.22(1): 1–10.doi:10.1093/nar/22.1.1.PMC307737.PMID8127644.
- ^Lauster R, Trautner TA, Noyer-Weidner M (March 1989). "Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains".Journal of Molecular Biology.206(2): 305–12.doi:10.1016/0022-2836(89)90480-4.PMID2716049.
- ^Cheng X (February 1995). "DNA modification by methyltransferases".Current Opinion in Structural Biology.5(1): 4–10.doi:10.1016/0959-440X(95)80003-J.PMID7773746.
- ^Cheng X, Kumar S, Posfai J, Pflugrath JW, Roberts RJ (July 1993). "Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine".Cell.74(2): 299–307.doi:10.1016/0092-8674(93)90421-L.PMID8343957.S2CID54238106.
- ^Oliveira PH, Fang G (May 2020)."Conserved DNA Methyltransferases: A Window into Fundamental Mechanisms of Epigenetic Regulation in Bacteria".Trends in Microbiology.29(1): 28–40.doi:10.1016/j.tim.2020.04.007.PMC7666040.PMID32417228.
- ^McClung CA, Nestler EJ (January 2008)."Neuroplasticity mediated by altered gene expression".Neuropsychopharmacology.33(1): 3–17.doi:10.1038/sj.npp.1301544.PMID17728700.S2CID18241370.
- ^abDuke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017)."Experience-dependent epigenomic reorganization in the hippocampus".Learn Mem.24(7): 278–288.doi:10.1101/lm.045112.117.PMC5473107.PMID28620075.
- ^"DNMT1".Gene Symbol Report.HUGO Gene Nomenclature Committee.Archived fromthe originalon 2012-10-06.Retrieved2012-09-27.
- ^Chen T, Ueda Y, Xie S, Li E (October 2002)."A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation".J Biol Chem.277(41): 38746–54.doi:10.1074/jbc.M205312200.PMID12138111.
- ^"DNMT3B".Gene Symbol Report.HUGO Gene Nomenclature Committee.Archived fromthe originalon 2012-11-20.Retrieved2012-09-27.
- ^Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, et al. (November 2016). "The DNA methyltransferase DNMT3C protects male germ cells from transposon activity".Science.354(6314): 909–912.Bibcode:2016Sci...354..909B.doi:10.1126/science.aah5143.PMID27856912.S2CID30907442.
- ^abManzo M, Wirz J, Ambrosi C, Villaseñor R, Roschitzki B, Baubec T (December 2017)."Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands".EMBO J.36(23): 3421–3434.doi:10.15252/embj.201797038.PMC5709737.PMID29074627.
- ^Rose NR, Klose RJ (December 2014)."Understanding the relationship between DNA methylation and histone lysine methylation".Biochim Biophys Acta.1839(12): 1362–72.doi:10.1016/j.bbagrm.2014.02.007.PMC4316174.PMID24560929.
- ^"DNMT3L".Gene Symbol Report.HUGO Gene Nomenclature Committee.Retrieved2012-09-27.
- ^Kho MR, Baker DJ, Laayoun A, Smith SS (January 1998). "Stalling of human DNA (cytosine-5) methyltransferase at single-strand conformers from a site of dynamic mutation".Journal of Molecular Biology.275(1): 67–79.doi:10.1006/jmbi.1997.1430.PMID9451440.
- ^Smith SS, Kaplan BE, Sowers LC, Newman EM (May 1992)."Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation".Proceedings of the National Academy of Sciences of the United States of America.89(10): 4744–8.Bibcode:1992PNAS...89.4744S.doi:10.1073/pnas.89.10.4744.PMC49160.PMID1584813.
- ^Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, et al. (January 2006). "De novo CpG island methylation in human cancer cells".Cancer Research.66(2): 682–92.doi:10.1158/0008-5472.CAN-05-1980.PMID16423997.
- ^Ting AH, Jair KW, Schuebel KE, Baylin SB (January 2006). "Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation".Cancer Research.66(2): 729–35.doi:10.1158/0008-5472.CAN-05-1537.PMID16424002.
- ^Li E, Bestor TH, Jaenisch R (June 1992). "Targeted mutation of the DNA methyltransferase gene results in embryonic lethality".Cell.69(6): 915–26.doi:10.1016/0092-8674(92)90611-F.PMID1606615.S2CID19879601.
- ^Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. (January 2006)."Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2".Science.311(5759): 395–8.Bibcode:2006Sci...311..395G.doi:10.1126/science.1120976.PMID16424344.S2CID39089541.
- ^"TRDMT1 tRNA aspartic acid methyltransferase 1 (Homo sapiens)".Entrez Gene.NCBI. 2010-11-01.Retrieved2010-11-07.
- ^Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G (October 2014)."Gene body methylation can alter gene expression and is a therapeutic target in cancer".Cancer Cell.26(4): 577–90.doi:10.1016/j.ccr.2014.07.028.PMC4224113.PMID25263941.
- ^abKaraca KG, Kupke J, Brito DV, Zeuch B, Thome C, Weichenhan D, Lutsik P, Plass C, Oliveira AM (January 2020)."Neuronal ensemble-specific DNA methylation strengthens engram stability".Nat Commun.11(1): 639.Bibcode:2020NatCo..11..639G.doi:10.1038/s41467-020-14498-4.PMC6994722.PMID32005851.
- ^Hegde M, Joshi MB (April 2021)."Comprehensive analysis of regulation of DNA methyltransferase isoforms in human breast tumors".J Cancer Res Clin Oncol.147(4): 937–971.doi:10.1007/s00432-021-03519-4.PMC7954751.PMID33604794.
- ^Oliveira AM, Hemstedt TJ, Bading H (July 2012). "Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities".Nat Neurosci.15(8): 1111–3.doi:10.1038/nn.3151.PMID22751036.S2CID10590208.
- ^Bernstein C (2022)."DNA Methylation and Establishing Memory".Epigenet Insights.15:25168657211072499.doi:10.1177/25168657211072499.PMC8793415.PMID35098021.
- ^Bayraktar G, Kreutz MR (2018)."The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders".Front Mol Neurosci.11:169.doi:10.3389/fnmol.2018.00169.PMC5975432.PMID29875631.
- ^Mack GS (December 2010). "To selectivity and beyond".Nature Biotechnology.28(12): 1259–66.doi:10.1038/nbt.1724.PMID21139608.S2CID11480326.
- ^"EC Approves Marketing Authorization Of DACOGEN For Acute Myeloid Leukemia".2012-09-28.Retrieved28 September2012.
Further reading[edit]
- Smith SS (1994).Biological implications of the mechanism of action of human DNA (cytosine-5)methyltransferase.Progress in Nucleic Acid Research and Molecular Biology. Vol. 49. pp. 65–111.doi:10.1016/s0079-6603(08)60048-3.ISBN9780125400497.PMID7863011.
- Pradhan S, Esteve PO (October 2003). "Mammalian DNA (cytosine-5) methyltransferases and their expression".Clinical Immunology.109(1): 6–16.doi:10.1016/S1521-6616(03)00204-3.PMID14585271.
- Goll MG, Bestor TH (2005). "Eukaryotic cytosine methyltransferases".Annual Review of Biochemistry.74:481–514.doi:10.1146/annurev.biochem.74.010904.153721.PMID15952895.S2CID32123961.
External links[edit]
- Information about DNA methyltransferases and DNA methylationat epigeneticstation.com
- Data for a DNA methyltransferase (DNMT) Antibody
- DNA+Modification+Methyltransferasesat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
