RNA

| Part of a series on |
| Genetics |
|---|
 |
Ribonucleic acid(RNA) is apolymericmolecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA anddeoxyribonucleic acid(DNA) arenucleic acids.The nucleic acids constitute one of the four majormacromoleculesessential for all known forms oflife.RNA is assembled as a chain ofnucleotides.Cellular organisms usemessenger RNA(mRNA) to convey genetic information (using thenitrogenous basesofguanine,uracil,adenine,andcytosine,denoted by the letters G, U, A, and C) that directs synthesis of specific proteins. Manyvirusesencode their genetic information using an RNAgenome.
Some RNA molecules play an active role within cells by catalyzing biological reactions, controllinggene expression,or sensing and communicating responses to cellular signals. One of these active processes isprotein synthesis,a universal function in which RNA molecules direct the synthesis of proteins onribosomes.This process usestransfer RNA(tRNA) molecules to deliveramino acidsto theribosome,whereribosomal RNA(rRNA) then links amino acids together to form coded proteins.
It has become widely accepted in science[1]that early in thehistory of life on Earth,prior to the evolution of DNA and possibly of protein-basedenzymesas well, an "RNA world"existed in which RNA served as both living organisms' storage method forgenetic information—a role fulfilled today by DNA, except in the case ofRNA viruses—and potentially performed catalytic functions in cells—a function performed today by protein enzymes, with the notable and important exception of the ribosome, which is aribozyme.
Comparison with DNA
[edit]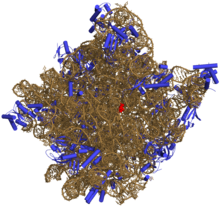
Like DNA, most biologically active RNAs, includingmRNA,tRNA,rRNA,snRNAs,and othernon-coding RNAs,contain self-complementary sequences that allow parts of the RNA to fold[6]and pair with itself to form double helices. Analysis of these RNAs has revealed that they are highly structured. Unlike DNA, their structures do not consist of long double helices, but rather collections of short helices packed together into structures akin to proteins.
In this fashion, RNAs can achieve chemicalcatalysis(like enzymes).[7]For instance, determination of the structure of the ribosome—an RNA-protein complex that catalyzes peptide bond formation—revealed that its active site is composed entirely of RNA.[8]
Structure
[edit]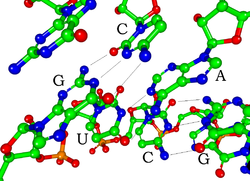
Eachnucleotidein RNA contains aribosesugar, with carbons numbered 1' through 5'. A base is attached to the 1' position, in general,adenine(A),cytosine(C),guanine(G), oruracil(U). Adenine and guanine arepurines,and cytosine and uracil arepyrimidines.Aphosphategroup is attached to the 3' position of one ribose and the 5' position of the next. The phosphate groups have a negative charge each, making RNA a charged molecule (polyanion). The bases formhydrogen bondsbetween cytosine and guanine, between adenine and uracil and between guanine and uracil.[9]However, other interactions are possible, such as a group of adenine bases binding to each other in a bulge,[10] or the GNRAtetraloopthat has a guanine–adenine base-pair.[9]
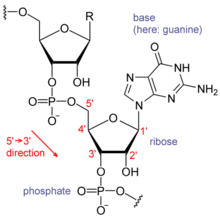
An important structural component of RNA that distinguishes it from DNA is the presence of ahydroxylgroup at the 2' position of theribose sugar.The presence of this functional group causes the helix to mostly take theA-form geometry,[11]although in single strand dinucleotide contexts, RNA can rarely also adopt the B-form most commonly observed in DNA.[12]The A-form geometry results in a very deep and narrow major groove and a shallow and wide minor groove.[13]A second consequence of the presence of the 2'-hydroxyl group is that in conformationally flexible regions of an RNA molecule (that is, not involved in formation of a double helix), it can chemically attack the adjacent phosphodiester bond to cleave the backbone.[14]
RNA is transcribed with only four bases (adenine, cytosine, guanine and uracil),[15]but these bases and attached sugars can be modified in numerous ways as the RNAs mature.Pseudouridine(Ψ), in which the linkage between uracil and ribose is changed from a C–N bond to a C–C bond, andribothymidine(T) are found in various places (the most notable ones being in the TΨC loop oftRNA).[16]Another notable modified base ishypoxanthine,a deaminated adenine base whosenucleosideis calledinosine(I). Inosine plays a key role in thewobble hypothesisof thegenetic code.[17]
There are more than 100 other naturally occurring modified nucleosides.[18]The greatest structural diversity of modifications can be found intRNA,[19]while pseudouridine and nucleosides with2'-O-methylriboseoften present in rRNA are the most common.[20]The specific roles of many of these modifications in RNA are not fully understood. However, it is notable that, in ribosomal RNA, many of the post-transcriptional modifications occur in highly functional regions, such as the peptidyl transferase center[21]and the subunit interface, implying that they are important for normal function.[22]
The functional form of single-stranded RNA molecules, just like proteins, frequently requires a specifictertiary structure.The scaffold for this structure is provided bysecondary structuralelements that are hydrogen bonds within the molecule. This leads to several recognizable "domains" of secondary structure likehairpin loops,bulges, andinternal loops.[23]In order to create, i.e., design, a RNA for any given secondary structure, two or three bases would not be enough, but four bases are enough.[24]This is likely why nature has "chosen" a four base alphabet: fewer than four would not allow the creation of all structures, while more than four bases are not necessary to do so. Since RNA is charged, metal ions such asMg2+are needed to stabilise many secondary andtertiary structures.[25]
The naturally occurringenantiomerof RNA isD-RNA composed ofD-ribonucleotides. All chirality centers are located in theD-ribose. By the use ofL-ribose or ratherL-ribonucleotides,L-RNA can be synthesized.L-RNA is much more stable against degradation byRNase.[26]
Like other structuredbiopolymerssuch as proteins, one can define topology of a folded RNA molecule. This is often done based on arrangement of intra-chain contacts within a folded RNA, termed ascircuit topology.
Synthesis
[edit]Synthesis of RNA is usually catalyzed by an enzyme—RNA polymerase—using DNA as a template, a process known astranscription.Initiation of transcription begins with the binding of the enzyme to apromotersequence in the DNA (usually found "upstream" of a gene). The DNA double helix is unwound by thehelicaseactivity of the enzyme. The enzyme then progresses along the template strand in the 3’ to 5’ direction, synthesizing a complementary RNA molecule with elongation occurring in the 5’ to 3’ direction. The DNA sequence also dictates where termination of RNA synthesis will occur.[27]
Primary transcriptRNAs are oftenmodifiedby enzymes after transcription. For example, apoly(A) tailand a5' capare added to eukaryoticpre-mRNAandintronsare removed by thespliceosome.
There are also a number ofRNA-dependent RNA polymerasesthat use RNA as their template for synthesis of a new strand of RNA. For instance, a number ofRNA viruses(such as poliovirus) use this type of enzyme to replicate their genetic material.[28]Also, RNA-dependent RNA polymerase is part of theRNA interferencepathway in many organisms.[29]
Types of RNA
[edit]Overview
[edit]
Messenger RNA (mRNA) is the RNA that carries information from DNA to theribosome,the sites of protein synthesis (translation) in the cell. The mRNA is a copy of DNA. The coding sequence of the mRNA determines theamino acidsequence in theproteinthat is produced.[30]However, many RNAs do not code for protein (about 97% of the transcriptional output is non-protein-coding in eukaryotes[31][32][33][34]).
These so-callednon-coding RNAs( "ncRNA" ) can be encoded by their own genes (RNA genes), but can also derive from mRNAintrons.[35]The most prominent examples of non-coding RNAs aretransfer RNA(tRNA) andribosomal RNA(rRNA), both of which are involved in the process of translation.[5]There are also non-coding RNAs involved in gene regulation,RNA processingand other roles. Certain RNAs are able tocatalysechemical reactions such as cutting andligatingother RNA molecules,[36]and the catalysis ofpeptide bondformation in theribosome;[8]these are known asribozymes.
In length
[edit]According to the length of RNA chain, RNA includessmall RNAand long RNA.[37]Usually,small RNAsare shorter than 200ntin length, and long RNAs are greater than 200ntlong.[38]Long RNAs, also called large RNAs, mainly includelong non-coding RNA(lncRNA) andmRNA.Small RNAs mainly include 5.8Sribosomal RNA(rRNA),5S rRNA,transfer RNA(tRNA),microRNA(miRNA),small interfering RNA(siRNA),small nucleolar RNA(snoRNAs),Piwi-interacting RNA(piRNA), tRNA-derived small RNA (tsRNA)[39]and small rDNA-derived RNA (srRNA).[40] There are certain exceptions as in the case of the5S rRNAof the members of the genusHalococcus(Archaea), which have an insertion, thus increasing its size.[41][42][43]
In translation
[edit]Messenger RNA(mRNA) carries information about a protein sequence to theribosomes,the protein synthesis factories in the cell. It iscodedso that every three nucleotides (acodon) corresponds to one amino acid. Ineukaryoticcells, once precursor mRNA (pre-mRNA) has been transcribed from DNA, it is processed to mature mRNA. This removes itsintrons—non-coding sections of the pre-mRNA. The mRNA is then exported from the nucleus to thecytoplasm,where it is bound to ribosomes andtranslatedinto its corresponding protein form with the help oftRNA.In prokaryotic cells, which do not have nucleus and cytoplasm compartments, mRNA can bind to ribosomes while it is being transcribed from DNA. After a certain amount of time, the message degrades into its component nucleotides with the assistance ofribonucleases.[30]
Transfer RNA(tRNA) is a small RNA chain of about 80nucleotidesthat transfers a specific amino acid to a growingpolypeptidechain at the ribosomal site of protein synthesis during translation. It has sites for amino acid attachment and ananticodonregion forcodonrecognition that binds to a specific sequence on the messenger RNA chain through hydrogen bonding.[35]

Ribosomal RNA(rRNA) is the catalytic component of the ribosomes. The rRNA is the component of the ribosome that hosts translation. Eukaryotic ribosomes contain four different rRNA molecules: 18S, 5.8S, 28S and 5S rRNA. Three of the rRNA molecules are synthesized in thenucleolus,and one is synthesized elsewhere. In the cytoplasm, ribosomal RNA and protein combine to form a nucleoprotein called a ribosome. The ribosome binds mRNA and carries out protein synthesis. Several ribosomes may be attached to a single mRNA at any time.[30]Nearly all the RNA found in a typical eukaryotic cell is rRNA.
Transfer-messenger RNA(tmRNA) is found in manybacteriaandplastids.It tags proteins encoded by mRNAs that lack stop codons for degradation and prevents the ribosome from stalling.[44]
Regulatory RNA
[edit]The earliest known regulators ofgene expressionwere proteins known asrepressorsandactivators– regulators with specific short binding sites withinenhancerregions near the genes to be regulated.[45]Later studies have shown that RNAs also regulate genes. There are several kinds of RNA-dependent processes in eukaryotes regulating the expression of genes at various points, such asRNAirepressing genespost-transcriptionally,long non-coding RNAsshutting down blocks ofchromatinepigenetically,andenhancer RNAsinducing increased gene expression.[46]Bacteria and archaeahave also been shown to use regulatory RNA systems such asbacterial small RNAsandCRISPR.[47]Fire and Mello were awarded the 2006Nobel Prize in Physiology or Medicinefor discoveringmicroRNAs(miRNAs), specific short RNA molecules that can base-pair with mRNAs.[48]
RNA interference by miRNAs
[edit]Post-transcriptional expression levels of many genes can be controlled byRNA interference,in whichmiRNAs,specific short RNA molecules, pair with mRNA regions and target them for degradation.[49]Thisantisense-based process involves steps that first process the RNA so that it canbase-pairwith a region of its target mRNAs. Once the base pairing occurs, other proteins direct the mRNA to be destroyed bynucleases.[46]
Long non-coding RNAs
[edit]Next to be linked to regulation wereXistand otherlong noncoding RNAsassociated withX chromosome inactivation.Their roles, at first mysterious, were shown byJeannie T. Leeand others to be thesilencingof blocks of chromatin via recruitment ofPolycombcomplex so that messenger RNA could not be transcribed from them.[50]Additional lncRNAs, currently defined as RNAs of more than 200 base pairs that do not appear to have coding potential,[51]have been found associated with regulation ofstem cellpluripotencyandcell division.[51]
Enhancer RNAs
[edit]The third major group of regulatory RNAs is calledenhancer RNAs.[51]It is not clear at present whether they are a unique category of RNAs of various lengths or constitute a distinct subset of lncRNAs. In any case, they are transcribed fromenhancers,which are known regulatory sites in the DNA near genes they regulate.[51][52]They up-regulate the transcription of the gene(s) under control of the enhancer from which they are transcribed.[51][53]
Regulatory RNA in prokaryotes
[edit]At first, regulatory RNA was thought to be a eukaryotic phenomenon, a part of the explanation for why so much more transcription in higher organisms was seen than had been predicted. But as soon as researchers began to look for possible RNA regulators in bacteria, they turned up there as well, termed as small RNA (sRNA).[54][47]Currently, the ubiquitous nature of systems of RNA regulation of genes has been discussed as support for theRNA Worldtheory.[46][55]There are indications that the enterobacterial sRNAs are involved in various cellular processes and seem to have significant role in stress responses such as membrane stress, starvation stress, phosphosugar stress and DNA damage. Also, it has been suggested that sRNAs have been evolved to have important role in stress responses because of their kinetic properties that allow for rapid response and stabilisation of the physiological state.[2]Bacterial small RNAsgenerally act viaantisensepairing with mRNA to down-regulate its translation, either by affecting stability or affecting cis-binding ability.[46]Riboswitcheshave also been discovered. They are cis-acting regulatory RNA sequences actingallosterically.They change shape when they bindmetabolitesso that they gain or lose the ability to bind chromatin to regulate expression of genes.[56][57]
Archaea also have systems of regulatory RNA.[58]The CRISPR system, recently being used to edit DNAin situ,acts via regulatory RNAs in archaea and bacteria to provide protection against virus invaders.[46][59]
In RNA processing
[edit]
Many RNAs are involved in modifying other RNAs. Intronsaresplicedout ofpre-mRNAbyspliceosomes,which contain severalsmall nuclear RNAs(snRNA),[5]or the introns can be ribozymes that are spliced by themselves.[60] RNA can also be altered by having its nucleotides modified to nucleotides other thanA,C,GandU. In eukaryotes, modifications of RNA nucleotides are in general directed bysmall nucleolar RNAs(snoRNA; 60–300 nt),[35]found in thenucleolusandcajal bodies.snoRNAs associate with enzymes and guide them to a spot on an RNA by basepairing to that RNA. These enzymes then perform the nucleotide modification. rRNAs and tRNAs are extensively modified, but snRNAs and mRNAs can also be the target of base modification.[61][62]RNA can also be methylated.[63][64]
RNA genomes
[edit]Like DNA, RNA can carry genetic information.RNA viruseshavegenomescomposed of RNA that encodes a number of proteins. The viral genome is replicated by some of those proteins, while other proteins protect the genome as the virus particle moves to a new host cell.Viroidsare another group of pathogens, but they consist only of RNA, do not encode any protein and are replicated by a host plant cell's polymerase.[65]
In reverse transcription
[edit]Reverse transcribing viruses replicate their genomes byreverse transcribingDNA copies from their RNA; these DNA copies are then transcribed to new RNA.Retrotransposonsalso spread by copying DNA and RNA from one another,[66]andtelomerasecontains anRNA that is used as templatefor building the ends ofeukaryotic chromosomes.[67]
Double-stranded RNA
[edit]
Double-stranded RNA(dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells, but with the replacement of thymine by uracil and the adding of one oxygen atom. dsRNA forms the genetic material of someviruses(double-stranded RNA viruses). Double-stranded RNA, such as viral RNA orsiRNA,can triggerRNA interferenceineukaryotes,as well asinterferonresponse invertebrates.[68][69][70][71]In eukaryotes, double-stranded RNA (dsRNA) plays a role in the activation of theinnate immune systemagainst viral infections.[72]
Circular RNA
[edit]In the late 1970s, it was shown that there is a single stranded covalently closed, i.e. circular form of RNA expressed throughout the animal and plant kingdom (seecircRNA).[73]circRNAs are thought to arise via a "back-splice" reaction where thespliceosomejoins a upstream 3' acceptor to a downstream 5' donor splice site. So far the function of circRNAs is largely unknown, although for few examples a microRNA sponging activity has been demonstrated.
Key discoveries in RNA biology
[edit]
Research on RNA has led to many important biological discoveries and numerousNobel Prizes.Nucleic acidswere discovered in 1868 byFriedrich Miescher,who called the material 'nuclein' since it was found in thenucleus.[74]It was later discovered that prokaryotic cells, which do not have a nucleus, also contain nucleic acids. The role of RNA in protein synthesis was suspected already in 1939.[75]Severo Ochoawon the 1959Nobel Prize in Medicine(shared withArthur Kornberg) after he discovered an enzyme that can synthesize RNA in the laboratory.[76]However, the enzyme discovered by Ochoa (polynucleotide phosphorylase) was later shown to be responsible for RNA degradation, not RNA synthesis. In 1956 Alex Rich and David Davies hybridized two separate strands of RNA to form the first crystal of RNA whose structure could be determined by X-ray crystallography.[77]
The sequence of the 77 nucleotides of a yeast tRNA was found byRobert W. Holleyin 1965,[78]winning Holley the1968 Nobel Prize in Medicine(shared withHar Gobind KhoranaandMarshall Nirenberg).
In the early 1970s,retrovirusesandreverse transcriptasewere discovered, showing for the first time that enzymes could copy RNA into DNA (the opposite of the usual route for transmission of genetic information). For this work,David Baltimore,Renato DulbeccoandHoward Teminwere awarded a Nobel Prize in 1975. In 1976,Walter Fiersand his team determined the first complete nucleotide sequence of an RNA virus genome, that ofbacteriophage MS2.[79]
In 1977,intronsandRNA splicingwere discovered in both mammalian viruses and in cellular genes, resulting in a 1993 Nobel toPhilip SharpandRichard Roberts. Catalytic RNA molecules (ribozymes) were discovered in the early 1980s, leading to a 1989 Nobel award toThomas CechandSidney Altman.In 1990, it was found inPetuniathat introduced genes can silence similar genes of the plant's own, now known to be a result ofRNA interference.[80][81]
At about the same time, 22 nt long RNAs, now calledmicroRNAs,were found to have a role in thedevelopmentofC. elegans.[82] Studies on RNA interference gleaned a Nobel Prize forAndrew FireandCraig Melloin 2006, and another Nobel was awarded for studies on the transcription of RNA toRoger Kornbergin the same year. The discovery of gene regulatory RNAs has led to attempts to develop drugs made of RNA, such assiRNA,to silence genes.[83]Adding to the Nobel prizes awarded for research on RNA in 2009 it was awarded for the elucidation of the atomic structure of the ribosome toVenki Ramakrishnan,Thomas A. Steitz,andAda Yonath.In 2023 theNobel Prize in Physiology or Medicinewas awarded toKatalin KarikóandDrew Weissmanfor their discoveries concerningmodified nucleosidesthat enabled the development of effective mRNA vaccines against COVID-19.[84][85][86]
Relevance for prebiotic chemistry and abiogenesis
[edit]In 1968,Carl Woesehypothesized that RNA might be catalytic and suggested that the earliest forms of life (self-replicating molecules) could have relied on RNA both to carry genetic information and to catalyze biochemical reactions—anRNA world.[87][88]In May 2022, scientists reported that they discovered RNA forms spontaneously on prebioticbasalt lava glasswhich is presumed to have been abundantly available on theearly Earth.[89][90]
In March 2015,DNAand RNAnucleobases,includinguracil,cytosineandthymine,were reportedly formed in the laboratory underouter spaceconditions, using starter chemicals, such aspyrimidine,anorganic compoundcommonly found inmeteorites.Pyrimidine,likepolycyclic aromatic hydrocarbons(PAHs), is one of the most carbon-rich compounds found in theUniverseand may have been formed inred giantsor ininterstellar dustand gas clouds.[91]In July 2022, astronomers reported the discovery of massive amounts ofprebiotic molecules,including possible RNA precursors, in theGalactic Centerof theMilky Way Galaxy.[92][93]
Medical applications
[edit]RNA, initially deemed unsuitable for therapeutic use due to its short half-life, has been proven to possess numerous therapeutic properties through advancements in stabilization chemistry. RNA molecules have potential therapeutic applications due to their ability to fold into complex conformations and binding proteins, nucleic acids, small molecules, and form catalytic centers.[94]RNA-based vaccines are thought to be a quicker way to obtain immunological resistance than the traditional approach of vaccines that rely on a killed or altered version of the pathogen, because it can take months or even years to grow and study a pathogen in order to determine which molecular parts to extract, inactivate, and use in a vaccine. Small molecules with conventional therapeutic properties can target RNA and DNA structures, thereby treating novel diseases. However, research on small molecules targeting RNA and approved drugs for human illness therapy is scarce. Ribavirin, branaplam, and ataluren are currently available medications that stabilize double-stranded RNA structures and control splicing in a variety of disorders.[95][96]
Protein-coding mRNAs have emerged as new therapeutic candidates, with RNA replacement being particularly beneficial for brief but torrent-like protein expression.[97]In vitro transcribed mRNAs (IVT-mRNA) have been used to deliver proteins for bone regeneration, pluripotency, and heart function in animal models.[98][99][100][101][102]SiRNAs, short RNA molecules, play a crucial role in innate defense against viruses and chromatin structure. They can be artificially introduced to silence specific genes, making them valuable for gene function studies, therapeutic target validation, and drug development.[97]
mRNA vaccineshave emerged as an important new class of vaccines, using mRNA to produce an immune response. Their first successful large-scale application came in the form ofCOVID-19 vaccinesduring theCOVID-19 pandemic.
See also
[edit]References
[edit]- ^Copley SD, Smith E, Markowitz HJ (December 2007). "The origin of the RNA world: co-evolution of genes and metabolism".Bioorganic Chemistry.35(6): 430–443.doi:10.1016/j.bioorg.2007.08.001.PMID17897696.
The proposal that life on Earth arose from an RNA World is widely accepted.
- ^ab"RNA: The Versatile Molecule".University of Utah.2015.
- ^"Nucleotides and Nucleic Acids"(PDF).University of California, Los Angeles.Archived fromthe original(PDF)on 2015-09-23.Retrieved2015-08-26.
- ^Shukla RN (2014).Analysis of Chromosomes.Agrotech Press.ISBN978-93-84568-17-7.[permanent dead link]
- ^abcBerg JM, Tymoczko JL, Stryer L (2002).Biochemistry(5th ed.). WH Freeman and Company. pp. 118–19, 781–808.ISBN978-0-7167-4684-3.OCLC179705944.
- ^Tinoco I, Bustamante C (October 1999). "How RNA folds".Journal of Molecular Biology.293(2): 271–81.doi:10.1006/jmbi.1999.3001.PMID10550208.
- ^Higgs PG (August 2000). "RNA secondary structure: physical and computational aspects".Quarterly Reviews of Biophysics.33(3): 199–253.doi:10.1017/S0033583500003620.PMID11191843.S2CID37230785.
- ^abNissen P, Hansen J, Ban N, Moore PB, Steitz TA (August 2000). "The structural basis of ribosome activity in peptide bond synthesis".Science.289(5481): 920–30.Bibcode:2000Sci...289..920N.doi:10.1126/science.289.5481.920.PMID10937990.
- ^abLee JC, Gutell RR (December 2004). "Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs".Journal of Molecular Biology.344(5): 1225–49.doi:10.1016/j.jmb.2004.09.072.PMID15561141.
- ^Barciszewski J, Frederic B, Clark C (1999).RNA biochemistry and biotechnology.Springer. pp. 73–87.ISBN978-0-7923-5862-6.OCLC52403776.
- ^Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR (April 1993). "The DNA strand in DNA.RNA hybrid duplexes is neither B-form nor A-form in solution".Biochemistry.32(16): 4207–15.doi:10.1021/bi00067a007.PMID7682844.
- ^Sedova A, Banavali NK (February 2016). "RNA approaches the B-form in stacked single strand dinucleotide contexts".Biopolymers.105(2): 65–82.doi:10.1002/bip.22750.PMID26443416.S2CID35949700.
- ^Hermann T, Patel DJ (March 2000)."RNA bulges as architectural and recognition motifs".Structure.8(3): R47–54.doi:10.1016/S0969-2126(00)00110-6.PMID10745015.
- ^Mikkola S, Stenman E, Nurmi K, Yousefi-Salakdeh E, Strömberg R, Lönnberg H (1999). "The mechanism of the metal ion promoted cleavage of RNA phosphodiester bonds involves a general acid catalysis by the metal aquo ion on the departure of the leaving group".Journal of the Chemical Society, Perkin Transactions 2(8): 1619–26.doi:10.1039/a903691a.
- ^Jankowski JA, Polak JM (1996).Clinical gene analysis and manipulation: Tools, techniques and troubleshooting.Cambridge University Press. p.14.ISBN978-0-521-47896-0.OCLC33838261.
- ^Yu Q, Morrow CD (May 2001)."Identification of critical elements in the tRNA acceptor stem and T(Psi)C loop necessary for human immunodeficiency virus type 1 infectivity".Journal of Virology.75(10): 4902–6.doi:10.1128/JVI.75.10.4902-4906.2001.PMC114245.PMID11312362.
- ^Elliott MS, Trewyn RW (February 1984)."Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine".The Journal of Biological Chemistry.259(4): 2407–10.doi:10.1016/S0021-9258(17)43367-9.PMID6365911.
- ^Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF (January 2011)."The RNA Modification Database, RNAMDB: 2011 update".Nucleic Acids Research.39(Database issue): D195-201.doi:10.1093/nar/gkq1028.PMC3013656.PMID21071406.
- ^Söll D, RajBhandary U (1995).TRNA: Structure, biosynthesis, and function.ASM Press. p. 165.ISBN978-1-55581-073-3.OCLC183036381.
- ^Kiss T (July 2001)."Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs".The EMBO Journal.20(14): 3617–22.doi:10.1093/emboj/20.14.3617.PMC125535.PMID11447102.
- ^Tirumalai MR, Rivas M, Tran Q, Fox GE (November 2021)."The Peptidyl Transferase Center: a Window to the Past".Microbiol Mol Biol Rev.85(4): e0010421.doi:10.1128/MMBR.00104-21.PMC8579967.PMID34756086.
- ^King TH, Liu B, McCully RR, Fournier MJ (February 2003)."Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center".Molecular Cell.11(2): 425–35.doi:10.1016/S1097-2765(03)00040-6.PMID12620230.
- ^Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH (May 2004)."Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure".Proceedings of the National Academy of Sciences of the United States of America.101(19): 7287–92.Bibcode:2004PNAS..101.7287M.doi:10.1073/pnas.0401799101.PMC409911.PMID15123812.
- ^Burghardt B, Hartmann AK (February 2007)."RNA secondary structure design".Physical Review E.75(2): 021920.arXiv:physics/0609135.Bibcode:2007PhRvE..75b1920B.doi:10.1103/PhysRevE.75.021920.PMID17358380.S2CID17574854.
- ^Tan ZJ, Chen SJ (July 2008)."Salt dependence of nucleic acid hairpin stability".Biophysical Journal.95(2): 738–52.Bibcode:2008BpJ....95..738T.doi:10.1529/biophysj.108.131524.PMC2440479.PMID18424500.
- ^Vater A, Klussmann S (January 2015)."Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer(®) therapeutics".Drug Discovery Today.20(1): 147–55.doi:10.1016/j.drudis.2014.09.004.PMID25236655.
- ^Nudler E, Gottesman ME (August 2002). "Transcription termination and anti-termination in E. coli".Genes to Cells.7(8): 755–68.doi:10.1046/j.1365-2443.2002.00563.x.PMID12167155.S2CID23191624.
- ^Hansen JL, Long AM, Schultz SC (August 1997)."Structure of the RNA-dependent RNA polymerase of poliovirus".Structure.5(8): 1109–22.doi:10.1016/S0969-2126(97)00261-X.PMID9309225.
- ^Ahlquist P (May 2002). "RNA-dependent RNA polymerases, viruses, and RNA silencing".Science.296(5571): 1270–73.Bibcode:2002Sci...296.1270A.doi:10.1126/science.1069132.PMID12016304.S2CID42526536.
- ^abcCooper GC, Hausman RE (2004).The Cell: A Molecular Approach(3rd ed.). Sinauer. pp. 261–76, 297, 339–44.ISBN978-0-87893-214-6.OCLC174924833.
- ^Mattick JS, Gagen MJ (September 2001)."The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms".Molecular Biology and Evolution.18(9): 1611–30.doi:10.1093/oxfordjournals.molbev.a003951.PMID11504843.
- ^Mattick JS (November 2001)."Non-coding RNAs: the architects of eukaryotic complexity".EMBO Reports.2(11): 986–91.doi:10.1093/embo-reports/kve230.PMC1084129.PMID11713189.
- ^Mattick JS (October 2003)."Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms"(PDF).BioEssays.25(10): 930–39.CiteSeerX10.1.1.476.7561.doi:10.1002/bies.10332.PMID14505360.Archived fromthe original(PDF)on 2009-03-06.
- ^Mattick JS (October 2004). "The hidden genetic program of complex organisms".Scientific American.291(4): 60–67.Bibcode:2004SciAm.291d..60M.doi:10.1038/scientificamerican1004-60.PMID15487671.[dead link]
- ^abcWirta W (2006).Mining the transcriptome – methods and applications.Stockholm: School of Biotechnology, Royal Institute of Technology.ISBN978-91-7178-436-0.OCLC185406288.
- ^Rossi JJ (July 2004)."Ribozyme diagnostics comes of age".Chemistry & Biology.11(7): 894–95.doi:10.1016/j.chembiol.2004.07.002.PMID15271347.
- ^Storz G (May 2002). "An expanding universe of noncoding RNAs".Science.296(5571): 1260–63.Bibcode:2002Sci...296.1260S.doi:10.1126/science.1072249.PMID12016301.S2CID35295924.
- ^Fatica A, Bozzoni I (January 2014)."Long non-coding RNAs: new players in cell differentiation and development".Nature Reviews Genetics.15(1): 7–21.doi:10.1038/nrg3606.PMID24296535.S2CID12295847.[permanent dead link]
- ^Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. (January 2016)."Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder"(PDF).Science.351(6271): 397–400.Bibcode:2016Sci...351..397C.doi:10.1126/science.aad7977.PMID26721680.S2CID21738301.
- ^Wei H, Zhou B, Zhang F, Tu Y, Hu Y, Zhang B, Zhai Q (2013)."Profiling and identification of small rDNA-derived RNAs and their potential biological functions".PLOS ONE.8(2): e56842.Bibcode:2013PLoSO...856842W.doi:10.1371/journal.pone.0056842.PMC3572043.PMID23418607.
- ^Luehrsen KR, Nicholson DE, Eubanks DC, Fox GE (1981). "An archaebacterial 5S rRNA contains a long insertion sequence".Nature.293(5835): 755–756.Bibcode:1981Natur.293..755L.doi:10.1038/293755a0.PMID6169998.S2CID4341755.
- ^Stan-Lotter H, McGenity TJ, Legat A, Denner EB, Glaser K, Stetter KO, Wanner G (1999)."Very similar strains of Halococcus salifodinae are found in geographically separated permo-triassic salt deposits".Microbiology.145(Pt 12): 3565–3574.doi:10.1099/00221287-145-12-3565.PMID10627054.
- ^Tirumalai MR, Kaelber JT, Park DR, Tran Q, Fox GE (August 2020)."Cryo-Electron Microscopy Visualization of a Large Insertion in the 5S ribosomal RNA of the Extremely Halophilic ArchaeonHalococcus morrhuae".FEBS Open Bio.10(10): 1938–1946.doi:10.1002/2211-5463.12962.PMC7530397.PMID32865340.
- ^Gueneau de Novoa P, Williams KP (January 2004)."The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts".Nucleic Acids Research.32(Database issue): D104–08.doi:10.1093/nar/gkh102.PMC308836.PMID14681369.
- ^Jacob F, Monod J (1961). "Genetic Regulatory Mechanisms in the Synthesis of Proteins".Journal of Molecular Biology.3(3): 318–56.doi:10.1016/s0022-2836(61)80072-7.PMID13718526.S2CID19804795.
- ^abcdeMorris K, Mattick J (2014)."The rise of regulatory RNA".Nature Reviews Genetics.15(6): 423–37.doi:10.1038/nrg3722.PMC4314111.PMID24776770.
- ^abGottesman S (2005). "Micros for microbes: non-coding regulatory RNAs in bacteria".Trends in Genetics.21(7): 399–404.doi:10.1016/j.tig.2005.05.008.PMID15913835.
- ^"The Nobel Prize in Physiology or Medicine 2006".Nobelprize.org.Nobel Media AB 2014. Web. 6 Aug 2018.http://www.nobelprize.org/nobel_prizes/medicine/laureates/2006
- ^Fire, et al. (1998)."Potent and Specific Genetic Interference by double stranded RNA in Ceanorhabditis elegans".Nature.391(6669): 806–11.Bibcode:1998Natur.391..806F.doi:10.1038/35888.PMID9486653.S2CID4355692.
- ^Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008)."Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome".Science.322(5902): 750–56.Bibcode:2008Sci...322..750Z.doi:10.1126/science.1163045.PMC2748911.PMID18974356.
- ^abcdeRinn JL, Chang HY (2012)."Genome regulation by long noncoding RNAs".Annu. Rev. Biochem.81:1–25.doi:10.1146/annurev-biochem-051410-092902.PMC3858397.PMID22663078.
- ^Taft RJ, Kaplan CD, Simons C, Mattick JS (2009)."Evolution, biogenesis and function of promoter- associated RNAs".Cell Cycle.8(15): 2332–38.doi:10.4161/cc.8.15.9154.PMID19597344.
- ^Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, et al. (2010)."'Long noncoding RNAs with enhancer-like function in human cells ".Cell.143(1): 46–58.doi:10.1016/j.cell.2010.09.001.PMC4108080.PMID20887892.
- ^EGH Wagner, P Romby. (2015). "Small RNAs in bacteria and archaea: who they are, what they do, and how they do it".Advances in genetics(Vol. 90, pp. 133–208).
- ^J.W. Nelson, R.R. Breaker (2017) "The lost language of the RNA World."Sci. Signal.10,eaam8812 1–11.
- ^Winklef WC (2005). "Riboswitches and the role of noncoding RNAs in bacterial metabolic control".Curr. Opin. Chem. Biol.9(6): 594–602.doi:10.1016/j.cbpa.2005.09.016.PMID16226486.
- ^Tucker BJ, Breaker RR (2005). "Riboswitches as versatile gene control elements".Curr. Opin. Struct. Biol.15(3): 342–48.doi:10.1016/j.sbi.2005.05.003.PMID15919195.
- ^Mojica FJ, Diez-Villasenor C, Soria E, Juez G (2000)."""Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria ".Mol. Microbiol.36(1): 244–46.doi:10.1046/j.1365-2958.2000.01838.x.PMID10760181.S2CID22216574.
- ^Brouns S, Jore MM, Lundgren M, Westra E, Slijkhuis R, Snijders A, Dickman M, Makarova K, Koonin E, Der Oost JV (2008)."Small CRISPR RNAs guide antiviral defense in prokaryotes".Science.321(5891): 960–64.Bibcode:2008Sci...321..960B.doi:10.1126/science.1159689.PMC5898235.PMID18703739.
- ^Steitz TA, Steitz JA (July 1993)."A general two-metal-ion mechanism for catalytic RNA".Proceedings of the National Academy of Sciences of the United States of America.90(14): 6498–502.Bibcode:1993PNAS...90.6498S.doi:10.1073/pnas.90.14.6498.PMC46959.PMID8341661.
- ^Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W (January 2007)."Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs".Nucleic Acids Research.35(Database issue): D183–87.doi:10.1093/nar/gkl873.PMC1669756.PMID17099227.
- ^Omer AD, Ziesche S, Decatur WA, Fournier MJ, Dennis PP (May 2003). "RNA-modifying machines in archaea".Molecular Microbiology.48(3): 617–29.doi:10.1046/j.1365-2958.2003.03483.x.PMID12694609.S2CID20326977.
- ^Cavaillé J, Nicoloso M, Bachellerie JP (October 1996)."Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides".Nature.383(6602): 732–35.Bibcode:1996Natur.383..732C.doi:10.1038/383732a0.PMID8878486.S2CID4334683.
- ^Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T (June 1996)."Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs".Cell.85(7): 1077–88.doi:10.1016/S0092-8674(00)81308-2.PMID8674114.S2CID10418885.
- ^Daròs JA, Elena SF, Flores R (June 2006)."Viroids: an Ariadne's thread into the RNA labyrinth".EMBO Reports.7(6): 593–98.doi:10.1038/sj.embor.7400706.PMC1479586.PMID16741503.
- ^Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (March 2004)."Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes".Genetics.166(3): 1437–50.doi:10.1534/genetics.166.3.1437.PMC1470764.PMID15082561.
- ^Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ (January 2008)."The telomerase database".Nucleic Acids Research.36(Database issue): D339–43.doi:10.1093/nar/gkm700.PMC2238860.PMID18073191.
- ^Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Hohn T, Pooggin MM (2006)."Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing".Nucleic Acids Research.34(21): 6233–46.doi:10.1093/nar/gkl886.PMC1669714.PMID17090584.
- ^Jana S, Chakraborty C, Nandi S, Deb JK (November 2004). "RNA interference: potential therapeutic targets".Applied Microbiology and Biotechnology.65(6): 649–57.doi:10.1007/s00253-004-1732-1.PMID15372214.S2CID20963666.
- ^Virol, J (May 2006)."Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses".Journal of Virology.80(10): 5059–5064.doi:10.1128/JVI.80.10.5059-5064.2006.PMC1472073.PMID16641297.
- ^Schultz U, Kaspers B, Staeheli P (May 2004). "The interferon system of non-mammalian vertebrates".Developmental and Comparative Immunology.28(5): 499–508.doi:10.1016/j.dci.2003.09.009.PMID15062646.
- ^Whitehead KA, Dahlman JE, Langer RS, Anderson DG (2011). "Silencing or stimulation? siRNA delivery and the immune system".Annual Review of Chemical and Biomolecular Engineering.2:77–96.doi:10.1146/annurev-chembioeng-061010-114133.PMID22432611.
- ^Hsu MT, Coca-Prados M (July 1979). "Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells".Nature.280(5720): 339–40.Bibcode:1979Natur.280..339H.doi:10.1038/280339a0.PMID460409.S2CID19968869.
- ^Dahm R (February 2005). "Friedrich Miescher and the discovery of DNA".Developmental Biology.278(2): 274–88.doi:10.1016/j.ydbio.2004.11.028.PMID15680349.
- ^Caspersson T, Schultz J (1939). "Pentose nucleotides in the cytoplasm of growing tissues".Nature.143(3623): 602–03.Bibcode:1939Natur.143..602C.doi:10.1038/143602c0.S2CID4140563.
- ^Ochoa S (1959)."Enzymatic synthesis of ribonucleic acid"(PDF).Nobel Lecture.
- ^Rich A, Davies D (1956). "A New Two-Stranded Helical Structure: Polyadenylic Acid and Polyuridylic Acid".Journal of the American Chemical Society.78(14): 3548–49.doi:10.1021/ja01595a086.
- ^Holley RW, et al. (March 1965). "Structure of a ribonucleic acid".Science.147(3664): 1462–65.Bibcode:1965Sci...147.1462H.doi:10.1126/science.147.3664.1462.PMID14263761.S2CID40989800.
- ^Fiers W, et al. (April 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene".Nature.260(5551): 500–07.Bibcode:1976Natur.260..500F.doi:10.1038/260500a0.PMID1264203.S2CID4289674.
- ^Napoli C, Lemieux C, Jorgensen R (April 1990)."Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans".The Plant Cell.2(4): 279–89.doi:10.1105/tpc.2.4.279.PMC159885.PMID12354959.
- ^Dafny-Yelin M, Chung SM, Frankman EL, Tzfira T (December 2007)."pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants".Plant Physiology.145(4): 1272–81.doi:10.1104/pp.107.106062.PMC2151715.PMID17766396.
- ^Ruvkun G (October 2001). "Molecular biology. Glimpses of a tiny RNA world".Science.294(5543): 797–99.doi:10.1126/science.1066315.PMID11679654.S2CID83506718.
- ^Fichou Y, Férec C (December 2006). "The potential of oligonucleotides for therapeutic applications".Trends in Biotechnology.24(12): 563–70.doi:10.1016/j.tibtech.2006.10.003.PMID17045686.
- ^"The Nobel Prize in Physiology or Medicine 2023".NobelPrize.org.Retrieved2023-10-03.
- ^"Hungarian and US scientists win Nobel for COVID-19 vaccine discoveries".Reuters.2023-10-02.Retrieved2023-10-03.
- ^"The Nobel Prize in Physiology or Medicine 2023".NobelPrize.org.Retrieved2023-10-03.
- ^Siebert S (2006)."Common sequence structure properties and stable regions in RNA secondary structures"(PDF).Dissertation, Albert-Ludwigs-Universität, Freiburg im Breisgau.p. 1. Archived fromthe original(PDF)on March 9, 2012.
- ^Szathmáry E (June 1999). "The origin of the genetic code: amino acids as cofactors in an RNA world".Trends in Genetics.15(6): 223–29.doi:10.1016/S0168-9525(99)01730-8.PMID10354582.
- ^Jerome, Craig A.; et al. (19 May 2022)."Catalytic Synthesis of Polyribonucleic Acid on Prebiotic Rock Glasses".Astrobiology.22(6): 629–636.Bibcode:2022AsBio..22..629J.doi:10.1089/ast.2022.0027.PMC9233534.PMID35588195.S2CID248917871.
- ^Foundation for Applied Molecular Evolution (3 June 2022)."Scientists announce a breakthrough in determining life's origin on Earth—and maybe Mars".Phys.org.Retrieved3 June2022.
- ^Marlaire R (3 March 2015)."NASA Ames Reproduces the Building Blocks of Life in Laboratory".NASA.Archived fromthe originalon 5 March 2015.Retrieved5 March2015.
- ^Starr, Michelle (8 July 2022)."Loads of Precursors For RNA Have Been Detected in The Center of Our Galaxy".ScienceAlert.Retrieved9 July2022.
- ^Rivilla, Victor M.; et al. (8 July 2022)."Molecular Precursors of the RNA-World in Space: New Nitriles in the G+0.693–0.027 Molecular Cloud".Frontiers in Astronomy and Space Sciences.9:876870.arXiv:2206.01053.Bibcode:2022FrASS...9.6870R.doi:10.3389/fspas.2022.876870.
- ^Cech, Thomas R.; Steitz, Joan A. (March 2014)."The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones".Cell.157(1): 77–94.doi:10.1016/j.cell.2014.03.008.ISSN0092-8674.PMID24679528.S2CID14852160.
- ^Palacino, James; Swalley, Susanne E; Song, Cheng; Cheung, Atwood K; Shu, Lei; Zhang, Xiaolu; Van Hoosear, Mailin; Shin, Youngah; Chin, Donovan N; Keller, Caroline Gubser; Beibel, Martin; Renaud, Nicole A; Smith, Thomas M; Salcius, Michael; Shi, Xiaoying (2015-06-01). "SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice".Nature Chemical Biology.11(7): 511–517.doi:10.1038/nchembio.1837.ISSN1552-4450.PMID26030728.
- ^Roy, Bijoyita; Friesen, Westley J.; Tomizawa, Yuki; Leszyk, John D.; Zhuo, Jin; Johnson, Briana; Dakka, Jumana; Trotta, Christopher R.; Xue, Xiaojiao; Mutyam, Venkateshwar; Keeling, Kim M.; Mobley, James A.; Rowe, Steven M.; Bedwell, David M.; Welch, Ellen M. (2016-10-04)."Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression".Proceedings of the National Academy of Sciences.113(44): 12508–12513.Bibcode:2016PNAS..11312508R.doi:10.1073/pnas.1605336113.ISSN0027-8424.PMC5098639.PMID27702906.
- ^abQadir, Muhammad Imran; Bukhat, Sherien; Rasul, Sumaira; Manzoor, Hamid; Manzoor, Majid (2019-09-03). "RNA therapeutics: Identification of novel targets leading to drug discovery".Journal of Cellular Biochemistry.121(2): 898–929.doi:10.1002/jcb.29364.ISSN0730-2312.PMID31478252.S2CID201806158.
- ^Balmayor, Elizabeth R.; Geiger, Johannes P.; Aneja, Manish K.; Berezhanskyy, Taras; Utzinger, Maximilian; Mykhaylyk, Olga; Rudolph, Carsten; Plank, Christian (May 2016). "Chemically modified RNA induces osteogenesis of stem cells and human tissue explants as well as accelerates bone healing in rats".Biomaterials.87:131–146.doi:10.1016/j.biomaterials.2016.02.018.ISSN0142-9612.PMID26923361.
- ^Plews, Jordan R.; Li, JianLiang; Jones, Mark; Moore, Harry D.; Mason, Chris; Andrews, Peter W.; Na, Jie (2010-12-30)."Activation of Pluripotency Genes in Human Fibroblast Cells by a Novel mRNA Based Approach".PLOS ONE.5(12): e14397.Bibcode:2010PLoSO...514397P.doi:10.1371/journal.pone.0014397.ISSN1932-6203.PMC3012685.PMID21209933.
- ^Preskey, David; Allison, Thomas F.; Jones, Mark; Mamchaoui, Kamel; Unger, Christian (May 2016). "Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts".Biochemical and Biophysical Research Communications.473(3): 743–751.doi:10.1016/j.bbrc.2015.09.102.ISSN0006-291X.PMID26449459.
- ^Warren, Luigi; Manos, Philip D.; Ahfeldt, Tim; Loh, Yuin-Han; Li, Hu; Lau, Frank; Ebina, Wataru; Mandal, Pankaj K.; Smith, Zachary D.; Meissner, Alexander; Daley, George Q.; Brack, Andrew S.; Collins, James J.; Cowan, Chad; Schlaeger, Thorsten M. (November 2010)."Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA".Cell Stem Cell.7(5): 618–630.doi:10.1016/j.stem.2010.08.012.ISSN1934-5909.PMC3656821.PMID20888316.
- ^Elangovan, Satheesh; Khorsand, Behnoush; Do, Anh-Vu; Hong, Liu; Dewerth, Alexander; Kormann, Michael; Ross, Ryan D.; Rick Sumner, D.; Allamargot, Chantal; Salem, Aliasger K. (November 2015)."Chemically modified RNA activated matrices enhance bone regeneration".Journal of Controlled Release.218:22–28.doi:10.1016/j.jconrel.2015.09.050.ISSN0168-3659.PMC4631704.PMID26415855.
External links
[edit]- RNA World websiteLink collection (structures, sequences, tools, journals)
- Nucleic Acid DatabaseImages of DNA, RNA, and complexes.
- Anna Marie Pyle's Seminar: RNA Structure, Function, and Recognition
