Gram-positive bacteria
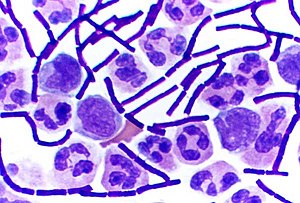
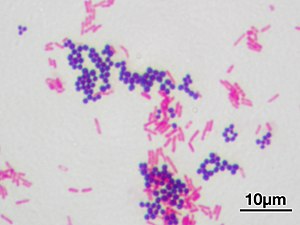
Inbacteriology,gram-positive bacteriaarebacteriathat give a positive result in theGram staintest, which is traditionally used to quickly classify bacteria into two broad categories according to their type ofcell wall.
The Gram stain is used by microbiologists to place bacteria into two main categories, Gram-positive (+) and Gram-negative (-). Gram-positive bacteria have a thick layer ofpeptidoglycanwithin the cell wall, and Gram-negative bacteria have a thin layer of peptidoglycan.
Gram-positive bacteria take up thecrystal violetstain used in the test, and then appear to be purple-coloured when seen through anoptical microscope.This is because the thick layer of peptidoglycan in the bacterial cell wall retains thestainafter it is washed away from the rest of the sample, in the decolorization stage of the test.
Conversely,gram-negative bacteriacannot retain the violet stain after the decolorization step;alcoholused in this stage degrades the outer membrane of gram-negative cells, making the cell wall more porous and incapable of retaining the crystal violet stain. Their peptidoglycan layer is much thinner and sandwiched between aninner cell membraneand abacterial outer membrane,causing them to take up thecounterstain(safraninorfuchsine) and appear red or pink.
Despite their thicker peptidoglycan layer, gram-positive bacteria are more receptive to certaincell wall–targetingantibioticsthan gram-negative bacteria, due to the absence of the outer membrane.[1]
Characteristics
[edit]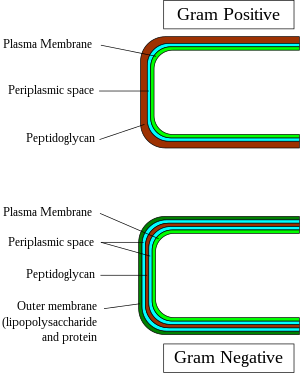

In general, the following characteristics are present in gram-positive bacteria:[2]
- Cytoplasmic lipid membrane
- Thickpeptidoglycanlayer
- Teichoic acidsand lipoids are present, forminglipoteichoic acids,which serve aschelatingagents, and also for certain types of adherence.
- Peptidoglycan chains are cross-linked to form rigid cell walls by a bacterial enzymeDD-transpeptidase.
- A much smaller volume ofperiplasmthan that in gram-negative bacteria.
Only some species have acapsule,usually consisting ofpolysaccharides.Also, only some species areflagellates,and when they do haveflagella,have only twobasal bodyrings to support them, whereas gram-negative have four. Both gram-positive and gram-negative bacteria commonly have a surface layer called anS-layer.In gram-positive bacteria, the S-layer is attached to the peptidoglycan layer. Gram-negative bacteria's S-layer is attached directly to theouter membrane.Specific to gram-positive bacteria is the presence ofteichoic acidsin the cell wall. Some of these are lipoteichoic acids, which have a lipid component in the cell membrane that can assist in anchoring the peptidoglycan.[3]
Classification
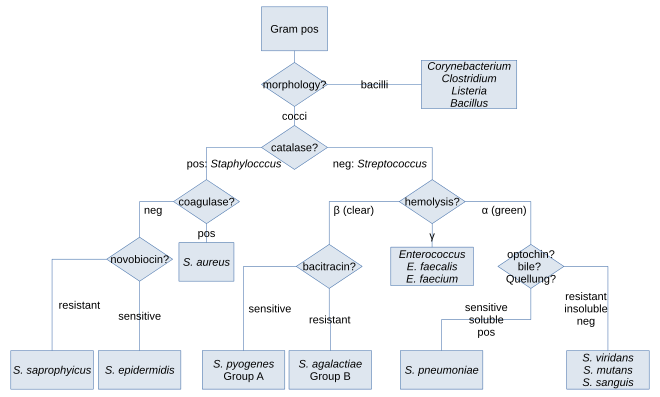
[edit]Along withcell shape,Gram staining is a rapid method used to differentiate bacterial species. Such staining, together with growth requirement and antibiotic susceptibility testing, and other macroscopic and physiologic tests, forms a basis for practical classification and subdivision of the bacteria (e.g., see figure and pre-1990 versions ofBergey's Manual of Systematic Bacteriology).[citation needed]
Historically,the kingdomMonerawas divided into fourdivisionsbased primarily on Gram staining:Bacillota(positive in staining),Gracilicutes(negative in staining),Mollicutes(neutral in staining) and Mendocutes (variable in staining).[4]Based on16S ribosomal RNAphylogenetic studies of the late microbiologistCarl Woeseand collaborators and colleagues at theUniversity of Illinois,themonophylyof the gram-positive bacteria was challenged,[5]with major implications for the therapeutic and general study of these organisms. Based onmolecular studiesof the 16S sequences, Woese recognised twelvebacterial phyla.Two of these were gram-positive and were divided on the proportion of theguanineandcytosinecontent in theirDNA.The high G + C phylum was made up of theActinobacteria,and the low G + C phylum contained theFirmicutes.[5]The Actinomycetota include theCorynebacterium,Mycobacterium,NocardiaandStreptomycesgenera. The (low G + C) Bacillota, have a 45–60% GC content, but this is lower than that of the Actinomycetota.[2]
Importance of the outer cell membrane in bacterial classification
[edit]It has been suggested that this section besplitout into another article titledGram stain.(Discuss)(November 2023) |

Although bacteria are traditionally divided into two main groups, gram-positive and gram-negative, based on their Gram stain retention property, this classification system is ambiguous as it refers to three distinct aspects (staining result, envelope organization, taxonomic group), which do not necessarily coalesce for some bacterial species.[6][7][8][9]The gram-positive and gram-negative staining response is also not a reliable characteristic as these two kinds of bacteria do not form phylogenetic coherent groups.[6]However, although Gram staining response is an empirical criterion, its basis lies in the marked differences in the ultrastructure and chemical composition of the bacterial cell wall, marked by the absence or presence of an outer lipid membrane.[6][10]
All gram-positive bacteria are bounded by a single-unit lipid membrane, and, in general, they contain a thick layer (20–80 nm) of peptidoglycan responsible for retaining the Gram stain. A number of other bacteria—that are bounded by a single membrane, but stain gram-negative due to either lack of the peptidoglycan layer, as in themycoplasmas,or their inability to retain the Gram stain because of their cell wall composition—also show close relationship to the gram-positive bacteria. For the bacterial cells bounded by a single cell membrane, the termmonoderm bacteriahas been proposed.[6][10]
In contrast to gram-positive bacteria, all typical gram-negative bacteria are bounded by a cytoplasmic membrane and an outer cell membrane; they contain only a thin layer of peptidoglycan (2–3 nm) between these membranes. The presence of inner and outer cell membranes defines a new compartment in these cells: theperiplasmic spaceor the periplasmic compartment. These bacteria have been designated asdiderm bacteria.[6][10]The distinction between the monoderm and diderm bacteria is supported by conserved signature indels in a number of important proteins (viz. DnaK, GroEL).[6][7][10][11]Of these two structurally distinct groups of bacteria, monoderms are indicated to be ancestral. Based upon a number of observations including that the gram-positive bacteria are the major producers of antibiotics and that, in general, gram-negative bacteria are resistant to them, it has been proposed that the outer cell membrane in gram-negative bacteria (diderms) has evolved as a protective mechanism againstantibioticselection pressure.[6][7][10][11]Some bacteria, such asDeinococcus,which stain gram-positive due to the presence of a thick peptidoglycan layer and also possess an outer cell membrane are suggested as intermediates in the transition between monoderm (gram-positive) and diderm (gram-negative) bacteria.[6][11]The diderm bacteria can also be further differentiated between simple diderms lacking lipopolysaccharide, the archetypical diderm bacteria where the outer cell membrane contains lipopolysaccharide, and the diderm bacteria where outer cell membrane is made up ofmycolic acid.[8][11][12]
Exceptions
[edit]In general, gram-positive bacteria are monoderms and have a singlelipid bilayerwhereas gram-negative bacteria are diderms and have two bilayers. Exceptions include:
- Some taxa lack peptidoglycan (such as the classMollicutes,some members of theRickettsiales,and the insect-endosymbionts of theEnterobacteriales) and aregram-indeterminate.
- TheDeinococcotahave gram-positive stains, although they are structurally similar to gram-negative bacteria with two layers.
- TheChloroflexotahave a single layer, yet (with some exceptions[13]) stain negative.[14]Two related phyla to the Chloroflexi, theTM7clade and the Ktedonobacteria, are also monoderms.[15][16]
Some Bacillota species are not gram-positive. The class Negativicutes, which includesSelenomonas,are diderm and stain gram-negative.[12]Additionally, a number of bacterial taxa (viz.Negativicutes,Fusobacteriota,Synergistota,andElusimicrobiota) that are either part of the phylum Bacillota or branch in its proximity are found to possess a diderm cell structure.[9][11][12]However, a conserved signature indel (CSI) in theHSP60(GroEL) protein distinguishes all traditional phyla of gram-negative bacteria (e.g.,Pseudomonadota,Aquificota,Chlamydiota,Bacteroidota,Chlorobiota,"Cyanobacteria",Fibrobacterota,Verrucomicrobiota,Planctomycetota,Spirochaetota,Acidobacteriota,etc.) from these other atypical diderm bacteria, as well as other phyla of monoderm bacteria (e.g.,Actinomycetota,Bacillota,Thermotogota,Chloroflexota,etc.).[11]The presence of this CSI in all sequenced species of conventional LPS (lipopolysaccharide)-containing gram-negative bacterial phyla provides evidence that these phyla of bacteria form a monophyletic clade and that no loss of the outer membrane from any species from this group has occurred.[11]
Pathogenicity
[edit]
In the classical sense, six gram-positive genera are typically pathogenic in humans. Two of these,StreptococcusandStaphylococcus,arecocci(sphere-shaped). The remaining organisms arebacilli(rod-shaped) and can be subdivided based on their ability to formspores.The non-spore formers areCorynebacteriumandListeria(a coccobacillus), whereasBacillusandClostridiumproduce spores.[17]The spore-forming bacteria can again be divided based on theirrespiration:Bacillusis afacultative anaerobe,whileClostridiumis anobligate anaerobe.[18]Also,Rathybacter,Leifsonia,andClavibacterare three gram-positive genera that cause plant disease. Gram-positive bacteria are capable of causing serious and sometimes fatalinfectionsin newborn infants.[19]Novel species of clinically relevant gram-positive bacteria also includeCatabacter hongkongensis,which is an emerging pathogen belonging toBacillota.[20]
Bacterial transformation
[edit]Transformationis one of three processes forhorizontal gene transfer,in which exogenous genetic material passes from a donor bacterium to a recipient bacterium, the other two processes beingconjugation(transfer ofgenetic materialbetween two bacterial cells in direct contact) andtransduction(injection of donor bacterial DNA by abacteriophagevirus into a recipient host bacterium).[21][22]In transformation, the genetic material passes through the intervening medium, and uptake is completely dependent on the recipient bacterium.[21]
As of 2014 about 80 species of bacteria were known to be capable of transformation, about evenly divided between gram-positive andgram-negative bacteria;the number might be an overestimate since several of the reports are supported by single papers.[21]Transformation among gram-positive bacteria has been studied in medically important species such asStreptococcus pneumoniae,Streptococcus mutans,Staphylococcus aureusandStreptococcus sanguinisand in gram-positive soil bacteriaBacillus subtilisandBacillus cereus.[23]
Orthographic note
[edit]The adjectivesgram-positiveandgram-negativederive from the surname ofHans Christian Gram;aseponymous adjectives,their initial letter can be either capitalGor lower-caseg,depending on whichstyle guide(e.g., that of theCDC), if any, governs the document being written.[24]
References
[edit]- ^Basic Biology (18 March 2016)."Bacteria".
- ^abMadigan, Michael T.; Martinko, John M. (2006).Brock Biology of Microorganisms(11th ed.). Pearson Prentice Hall.ISBN978-0131443297.
- ^Brown, Stephanie; Santa Maria, John P.; Walker, Suzanne (2013-09-08)."Wall Teichoic Acids of Gram-Positive Bacteria".Annual Review of Microbiology.67(1): 313–336.doi:10.1146/annurev-micro-092412-155620.ISSN0066-4227.PMC3883102.PMID24024634.
- ^Gibbons, N.E.; Murray, R.G.E. (1978)."Proposals Concerning the Higher Taxa of Bacteria".International Journal of Systematic and Evolutionary Microbiology.28(1): 1–6.doi:10.1099/00207713-28-1-1.
- ^abWoese, C.R. (1987)."Bacterial evolution".Microbiological Reviews.51(2): 221–271.doi:10.1128/MMBR.51.2.221-271.1987.PMC373105.PMID2439888.
- ^abcdefghGupta, R.S. (1998)."Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria and eukaryotes".Microbiology and Molecular Biology Reviews.62(4): 1435–1491.doi:10.1128/MMBR.62.4.1435-1491.1998.PMC98952.PMID9841678.
- ^abcGupta, R.S. (2000)."The natural evolutionary relationships among prokaryotes"(PDF).Critical Reviews in Microbiology.26(2): 111–131.CiteSeerX10.1.1.496.1356.doi:10.1080/10408410091154219.PMID10890353.S2CID30541897.Archived(PDF)from the original on 2013-06-25.
- ^abDesvaux, M.; Hébraud, M.; Talon, R.; Henderson, I.R. (2009). "Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue".Trends in Microbiology.17(4): 139–145.doi:10.1016/j.tim.2009.01.004.PMID19299134.
- ^abSutcliffe, I.C. (2010). "A phylum level perspective on bacterial cell envelope architecture".Trends in Microbiology.18(10): 464–470.doi:10.1016/j.tim.2010.06.005.PMID20637628.
- ^abcdeGupta, R.S. (1998). "What are archaebacteria: life's third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms".Molecular Microbiology.29(3): 695–707.doi:10.1046/j.1365-2958.1998.00978.x.PMID9723910.S2CID41206658.
- ^abcdefgGupta, R.S. (2011)."Origin of diderm (gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes".Antonie van Leeuwenhoek.100(2): 171–182.doi:10.1007/s10482-011-9616-8.PMC3133647.PMID21717204.
- ^abcMarchandin, H.; Teyssier, C.; Campos, J.; Jean-Pierre, H.; Roger, F.; Gay, B.; Carlier, J.-P.; Jumas-Bilak, E. (2009)."Negativicoccus succinicivorans gen. Nov., sp. Nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. In the bacterial phylum Firmicutes".International Journal of Systematic and Evolutionary Microbiology.60(6): 1271–1279.doi:10.1099/ijs.0.013102-0.PMID19667386.
- ^Yabe, S.; Aiba, Y.; Sakai, Y.; Hazaka, M.; Yokota, A. (2010)."Thermogemmatispora onikobensisgen. nov., sp. nov. AndThermogemmatispora foliorumsp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales ord. Nov. Within the class Ktedonobacteria ".International Journal of Systematic and Evolutionary Microbiology.61(4): 903–910.doi:10.1099/ijs.0.024877-0.PMID20495028.
- ^Sutcliffe, I.C. (2011). "Cell envelope architecture in the Chloroflexi: A shifting frontline in a phylogenetic turf war".Environmental Microbiology.13(2): 279–282.Bibcode:2011EnvMi..13..279S.doi:10.1111/j.1462-2920.2010.02339.x.PMID20860732.
- ^Hugenholtz, P.; Tyson, G.W.; Webb, R.I.; Wagner, A.M.; Blackall, L.L. (2001)."Investigation of Candidate Division TM7, a Recently Recognized Major Lineage of the Domain Bacteria with No Known Pure-Culture Representatives".Applied and Environmental Microbiology.67(1): 411–419.Bibcode:2001ApEnM..67..411H.doi:10.1128/AEM.67.1.411-419.2001.PMC92593.PMID11133473.
- ^Cavaletti, L.; Monciardini, P.; Bamonte, R.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. (2006)."New Lineage of Filamentous, Spore-Forming, Gram-Positive Bacteria from Soil".Applied and Environmental Microbiology.72(6): 4360–4369.Bibcode:2006ApEnM..72.4360C.doi:10.1128/AEM.00132-06.PMC1489649.PMID16751552.
- ^Gladwin, Mark; Trattler, Bill (2007).Clinical Microbiology Made Ridiculously Simple.Miami, FL: MedMaster. pp. 4–5.ISBN978-0-940780-81-1.
- ^Sahebnasagh, R.; Saderi, H.; Owlia, P. (4–7 September 2011).Detection of methicillin-resistantStaphylococcus aureusstrains from clinical samples in Tehran by detection of themecAandnucgenes.The First Iranian International Congress of Medical Bacteriology. Tabriz, Iran.
- ^MacDonald, Mhairi (2015).Avery's Neonatology: Pathophysiology and Management of the Newborn.Philadelphia, PA: Wolters Kluwer.ISBN9781451192681.Access provided by the University of Pittsburgh.
- ^Lau, S.K.P.; McNabb, A.; Woo, G.K.S.; Hoang, L.; Fung, A.M.Y.; Chung, L.M.W.; Woo, P.C.Y.; Yuen, K.-Y. (22 November 2006)."Catabacter hongkongensis gen. nov., sp. nov., Isolated from Blood Cultures of Patients from Hong Kong and Canada".Journal of Clinical Microbiology.45(2): 395–401.doi:10.1128/jcm.01831-06.ISSN0095-1137.PMC1829005.PMID17122022.
- ^abcJohnston, C.; Martin, B.; Fichant, G.; Polard, P; Claverys, J.P. (2014). "Bacterial transformation: distribution, shared mechanisms and divergent control".Nature Reviews. Microbiology.12(3): 181–196.doi:10.1038/nrmicro3199.PMID24509783.S2CID23559881.
- ^Korotetskiy I, Shilov S, Kuznetsova T, Kerimzhanova B, Korotetskaya N, Ivanova L, Zubenko N, Parenova R, Reva O (2023)."Analysis of Whole-Genome Sequences of Pathogenic Gram-Positive and Gram-Negative Isolates from the Same Hospital Environment to Investigate Common Evolutionary Trends Associated with Horizontal Gene Exchange, Mutations and DNA Methylation Patterning".Microorganisms.11(2): 323.doi:10.3390/microorganisms11020323.PMC9961978.PMID36838287.
- ^Michod, R.E.; Bernstein, H.; Nedelcu, A.M. (2008). "Adaptive value of sex in microbial pathogens".Infection, Genetics and Evolution.8(3): 267–285.Bibcode:2008InfGE...8..267M.doi:10.1016/j.meegid.2008.01.002.PMID18295550.
- ^"Emerging Infectious DiseasesJournal Style Guide ".CDC.gov.Centers for Disease Control and Prevention.
External links
[edit] This article incorporatespublic domain materialfromScience Primer.NCBI.Archived fromthe originalon 2009-12-08.
This article incorporatespublic domain materialfromScience Primer.NCBI.Archived fromthe originalon 2009-12-08.- 3D structures of proteins associated with plasma membrane of gram-positive bacteria
- 3D structures of proteins associated with outer membrane of gram-positive bacteria
