HLA-A3
This articlemay be too technical for most readers to understand.(May 2017) |
| HLA-A3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I,Acell surface antigen) | ||||||||||
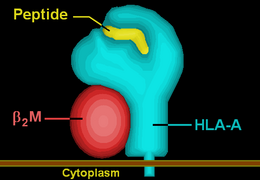 HLA-A3 | ||||||||||
| About | ||||||||||
| Protein | transmembrane receptor/ligand | |||||||||
| Structure | αβheterodimer | |||||||||
| Subunits | HLA-A*03--,β2-microglobulin | |||||||||
| Older names | HL-A3 | |||||||||
| Subtypes | ||||||||||
| ||||||||||
| Rare alleles | ||||||||||
| ||||||||||
| Alleles link-out toIMGT/HLAdatabase atEBI | ||||||||||
HLA-A3(A3) is ahuman leukocyte antigenserotypewithinHLA-Aserotype group. The serotype is determined by the antibody recognition of α3subset of HLA-A α-chains. For A3, the alpha, "A", chain are encoded by the HLA-A*03allele group and the β-chain are encoded byB2Mlocus.[1]This group currently is dominated by A*03:01. A3 and A*03are almost synonymous in meaning. A3 is more common in Europe, it is part of the longest known multigenehaplotype,A3~B7~DR15~DQ6.[2]
Serotype
[edit]| A*03 | A3 | Sample |
| allele | % | size (N) |
| *03:01 | 99 | 3504 |
| *03:02 | 78 | 342 |
| *03:05 | 20 | 5 |
A3 is primarily composed of A*03:01 and *03:02 which serotype well with anti-A3 antibodies. There are 26 non-synonymous variants of A*03, 4 nulls, and 22 protein variants.
Associated diseases
[edit]A3 serotype is a secondary risk factor formyasthenia gravis[4]and lower CD8+levels inhemochromatosispatients.[5][6]The HFE (Hemochromatosis) locus lies between A3 and B7 within the A3~DQ6 superhaplotype.[7]
In HIV
[edit]HLA-A3 selects HIV evolution for a mutation Gag KK9 epitope and results in a rapid decline in the CD8 T-cell response. CD8 T-cells are responsible for quickly killing HIV infected CD4+ cells.[8]This type of evolved response may not be specific for HLA-A3 and since HIV is capable of adapting quickly in situ to selective factors.
Alleles
[edit]| Study population | Freq. (in %)[9] |
|---|---|
| Finland | 25.0 |
| India Tamil Nadu Nadar | 20.5 |
| Czech Republic | 18.9 |
| Belgium | 17.1 |
| Ireland Northern | 14.3 |
| Australia New South Wales | 13.8 |
| Georgia Svaneti Svans | 13.8 |
| Georgia Tbilisi Georgian… | 13.8 |
| Pakistan Burusho | 13.0 |
| Cape Verde Northwestern I… | 12.0 |
| Ireland South | 11.6 |
| Romanian | 11.6 |
| Croatia | 11.0 |
| USA Caucasians (3) | 10.8 |
| Italy North (1) | 9.7 |
| USA African Americans (2) | 9.4 |
| India Mumbai Marathas | 9.3 |
| India West Bhils | 9.0 |
| Portugal Centre | 9.0 |
| Pakistan Pathan | 8.7 |
| Cameroon Beti | 8.6 |
| Cameroon Bamileke | 7.8 |
| Pakistan Kalash | 7.5 |
| Saudi Arabia Guraiat and… | 7.5 |
| USA Hispanic | 7.3 |
| India North Delhi | 7.2 |
| Madeira | 7.0 |
| Russia Tuva (2) | 6.9 |
| Tunisia | 6.7 |
| USA North American Native… | 6.7 |
| Cape Verde Southeastern I… | 6.5 |
| Azores Santa Maria and Sa… | 6.4 |
| Pakistan Baloch | 6.3 |
| Guinea Bissau | 6.2 |
| Israel Arab Druse | 6.0 |
| South African Natal Zulu | 6.0 |
| USA South Texas Hispanics | 6.0 |
| India North Hindus | 5.8 |
| Zambia Lusaka | 5.8 |
| Iran Baloch | 5.6 |
| Uganda Kampala | 5.5 |
| Jordan Amman | 5.2 |
| Kenya | 5.2 |
| Brazil | 5.1 |
| Oman | 5.1 |
| Cameroon Pygmy Baka | 5.0 |
| Georgia Tbilisi Kurds | 5.0 |
| Sudanese | 4.8 |
| Bulgaria | 4.6 |
| Pakistan Brahui | 4.6 |
| China Beijing | 4.5 |
| Australia Indig. Cape Yor… | 4.4 |
| China Inner Mongolia | 4.4 |
| Mali Bandiagara | 4.4 |
| Pakistan Sindhi | 4.4 |
| China Qinghai Hui | 4.1 |
| Zimbabwe Harare Shona | 4.0 |
| Cameroon Sawa | 3.8 |
| Senegal Niokholo Mandenka | 3.8 |
| Kenya Luo | 3.6 |
| Morocco Nador Metalsa Cla… | 3.4 |
| Kenya Nandi | 3.1 |
| Mex. Guadalajara Mestizos… | 3.1 |
| China Guangzhou | 2.9 |
| China North Han | 2.9 |
| Mongolia Buriat | 2.9 |
| China Tibetans | 2.5 |
| China Yunnan Nu | 2.5 |
| Mexico Mestizos | 2.4 |
| American Samoa | 2.0 |
| Singapore Javanese Indone… | 2.0 |
| Allele frequenciespresented, only | |
| Study population | Freq. (in %)[9] |
|---|---|
| Georgia Tbilisi Kurds | 8.3 |
| Morocco Nador Metalsa Cla… | 4.1 |
| Pakistan Karachi Parsi | 2.8 |
| Israel Arab Druse | 2.5 |
| India New Delhi | 2.3 |
| Sudanese | 2.3 |
| India North Delhi | 2.2 |
| India West Coast Parsis | 2.0 |
| Portugal Centre | 2.0 |
| India Andhra Pradesh Goll… | 1.7 |
| Pakistan Kalash | 1.7 |
| Pakistan Baloch | 1.6 |
| Georgia Tbilisi Georgian… | 1.4 |
| Saudi Arabia Guraiat and… | 1.4 |
| Oman | 1.3 |
| Pakistan Sindhi | 1.3 |
| India North Hindus | 1.0 |
| Pakistan Pathan | 1.0 |
| South Africa Natal Tamil | 1.0 |
| USA South Texas Hispanics | 0.8 |
| Allele frequenciespresented, only | |
Associated diseases
[edit]A*03:01 modulates increased risk formultiple sclerosis[10]
A3~B haplotypes
[edit]A3-B7 is part of the A3~DQ2 superhaplotype
A3-B8 (Romania, svanS)
A3~B35 (Bulgaria, Croatia, E. Black Sea)
A3~B55 (E. Black Sea)
A3~Cw7~B7
[edit]A3~B7 is bimodal in frequency in Europe with one node in Ireland and the other in Switzerland, relatively speaking Switzerland appears to be higher. A3~Cw7~B7 is one of the most common multigene haplotypes in the western world, particularly in Central and Eastern Europe.
A*03:01~ C*07:02~ B*07:02~ DRB1*15:01~ DQA1*01:02~ DQB1*06:02
References
[edit]- ^Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin".Tissue Antigens.11(2): 96–112.doi:10.1111/j.1399-0039.1978.tb01233.x.PMID77067.
- ^Horton R, Gibson R, Coggill P, et al. (January 2008)."Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project".Immunogenetics.60(1): 1–18.doi:10.1007/s00251-007-0262-2.PMC2206249.PMID18193213.
- ^Allele Query FormIMGT/HLA-European Bioinformatics Institute
- ^Machens A, Löliger C, Pichlmeier U, Emskötter T, Busch C, Izbicki J (1999). "Correlation of thymic pathology with HLA in myasthenia gravis".Clinical Immunology.91(3): 296–301.doi:10.1006/clim.1999.4710.PMID10370374.
- ^Barton J, Wiener H, Acton R, Go R (2005). "HLA haplotype A*03-B*07 in hemochromatosis probands with HFE C282Y homozygosity: frequency disparity in men and women and lack of association with severity of iron overload".Blood Cells Mol Dis.34(1): 38–47.doi:10.1016/j.bcmd.2004.08.022.PMID15607698.
- ^Cruz E, Vieira J, Almeida S, Lacerda R, Gartner A, Cardoso C, Alves H, Porto G (2006)."A study of 82 extended HLA haplotypes in HFE-C282Y homozygous hemochromatosis subjects: relationship to the genetic control of CD8+ T-lymphocyte numbers and severity of iron overload".BMC Med Genet.7:16.doi:10.1186/1471-2350-7-16.PMC1413516.PMID16509978.
- ^Olsson KS, Ritter B, Hansson N, Chowdhury RR (July 2008)."HLA haplotype map of river valley populations with hemochromatosis traced through five centuries in Central Sweden".Eur. J. Haematol.81(1): 36–46.doi:10.1111/j.1600-0609.2008.01078.x.PMID18363869.S2CID30689398.
- ^Allen TM, Altfeld M, Yu XG, et al. (July 2004)."Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection".J. Virol.78(13): 7069–78.doi:10.1128/JVI.78.13.7069-7078.2004.PMC421658.PMID15194783.
- ^abMiddleton, D.; Menchaca, L.; Rood, H.; Komerofsky, R. (2003). "New allele frequency database: http://www.allelefrequencies.net".Tissue Antigens.61(5): 403–407.doi:10.1034/j.1399-0039.2003.00062.x.PMID12753660.
- ^Fogdell-Hahn A, Ligers A, Grønning M, Hillert J, Olerup O (2000). "Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease".Tissue Antigens.55(2): 140–8.doi:10.1034/j.1399-0039.2000.550205.x.PMID10746785.
