Asgard (archaea)
| Asgard | |
|---|---|
| Scientific classification | |
| Domain: | Archaea |
| Kingdom: | Proteoarchaeota |
| Superphylum: | Asgard Katarzyna Zaremba-Niedzwiedzka,et al.2017 |
| Phyla | |
|
see text | |
| |
| Synonyms | |
| |
AsgardorAsgardarchaeota[2]is a proposedsuperphylumconsisting of a group ofarchaeathat contain eukaryotic signature proteins.[3]It appears that theeukaryotes,thedomainthat contains theanimals,plants,andfungi,emerged within the Asgard,[4]in a branch containing the Heimdallarchaeota.[5]This supports thetwo-domain systemof classification over thethree-domain system.[6][7]
Discovery and nomenclature
[edit]In the summer of 2010, sediments were analysed from a gravitycoretaken in the rift valley on the Knipovich ridge in the Arctic Ocean, near theLoki's Castlehydrothermal ventsite. Specific sediment horizons previously shown to contain high abundances of novel archaeal lineages were subjected tometagenomic analysis.[8][9]In 2015, anUppsala University-led team proposed theLokiarchaeotaphylum based onphylogeneticanalyses using a set ofhighly conservedprotein-coding genes.[10]The group was named for the shape-shifting Norse godLoki,in an allusion to the hydrothermal vent complex from which the first genome sample originated.[11]The Loki of mythology has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies",[12]analogous to the role of Lokiarchaeota in the debates about the origin of eukaryotes.[10][13]
In 2016, aUniversity of Texas-led team discoveredThorarchaeotafrom samples taken from theWhite Oak Riverin North Carolina, named in reference toThor,another Norse god.[14]Samples from Loki's Castle,Yellowstone National Park,Aarhus Bay,an aquifer near theColorado River,New Zealand'sRadiata Pool,hydrothermal vents nearTaketomi Island,Japan, and theWhite Oak Riverestuary in the United States contained Odinarchaeota and Heimdallarchaeota;[3]following the Norse deity naming convention, these groups were named forOdinandHeimdallrespectively. Researchers therefore named the superphylum containing these microbes "Asgard",after the home of the gods in Norse mythology.[3]Two Lokiarchaeota specimens have been cultured, enabling a detailed insight into their morphology.[15]
Description
[edit]
Proteins
[edit]Asgard members encode many eukaryotic signature proteins, including novelGTPases,membrane-remodelling proteins likeESCRTandSNF7,aubiquitinmodifier system, andN-glycosylationpathway homologs.[3]
Asgard archaeons have a regulatedactincytoskeleton,and theprofilinsandgelsolinsthey use can interact with eukaryotic actins.[16][17]In addition, Asgard archaeatubulinfrom hydrothermal-living Odinarchaeota (OdinTubulin) was identified as a genuine tubulin. OdinTubulin forms protomers and protofilaments most similar to eukaryotic microtubules, yet assembles into ring systems more similar toFtsZ,indicating that OdinTubulin may represent an evolution intermediate between FtsZ andmicrotubule-forming tubulins.[18]They also seem to form vesicles undercryogenic electron microscopy.Some may have aPKD domainS-layer.[19]They also share the three-way ES39 expansion inLSU rRNAwith eukaryotes.[20]Gene clusters or operons encoding ribosomal proteins are often less conserved in their organization in the Asgard group than in other Archaea, suggesting that the order of ribosomal protein coding genes may follow the phylogeny.[21]
Metabolism
[edit]-
Metabolic pathways of Asgard archaea, varying by phyla[22]
-
Metabolic pathways of Asgard archaea, varying by environment[22]
Asgard archaea are generallyobligate anaerobes,though Kariarchaeota, Gerdarchaeota and Hodarchaeota may befacultative aerobes.[23]They have aWood–Ljungdahl pathwayand performglycolysis.Members can beautotrophs,heterotrophs,orphototrophsusingheliorhodopsin.[22]One member,CandidatusPrometheoarchaeum syntrophicum,issyntrophicwith a sulfur-reducing proteobacteria and amethanogenicarchaea.[19]
TheRuBisCOthey have is not carbon-fixing, but likely used for nucleoside salvaging.[22]
Ecology
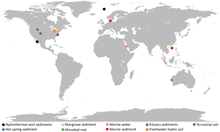
[edit]Asgard are widely distributed around the world, both geographically and by habitat. Many of the known clades are restricted to sediments, whereas Lokiarchaeota, Thorarchaeota and another clade occupy many different habitats. Salinity and depth are important ecological drivers for most Asgard archaea. Other habitats include the bodies of animals, the rhizosphere of plants, non-saline sediments and soils, the sea surface, and freshwater. In addition, Asgard are associated with several other microorganisms.[24]
Eukaryote-like features in subdivisions
[edit]The phylum Heimdallarchaeota was found in 2017 to have N-terminal corehistone tails,a feature previously thought to be exclusively eukaryotic. Two other archaeal phyla, both outside of Asgard, were found to also have tails in 2018.[25]
In January 2020, scientists foundCandidatusPrometheoarchaeum syntrophicum,a member of the Lokiarcheota, engaging incross-feedingwith two bacterial species. Drawing an analogy tosymbiogenesis,they consider this relationship a possible link between the simpleprokaryoticmicroorganisms and the complexeukaryoticmicroorganisms occurring approximately two billion years ago.[26][19]
Phylogeny
[edit]The phylogenetic relationships of the Asgard archaea have been studied by several teams in the 21st century.[5][4][27][23]Varying results have been obtained, for instance using 53 marker proteins from theGenome Taxonomy Database.[28][29][30]In 2023, Eme, Tamarit, Caceres and colleagues reported that the Eukaryota are deep within Asgard, as sister of Hodarchaeales within the Heimdallarchaeia.[31]
Taxonomy
[edit]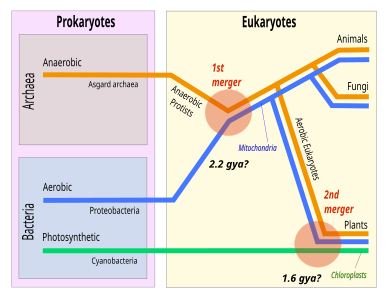
In the depicted scenario, the Eukaryota are deep in the tree of Asgard. A favored scenario is syntrophy, where one organism depends on the feeding of the other. Anα-proteobacteriumwas incorporated to become themitochondrion.[33]In culture, extant Asgard archaea form various syntrophic dependencies.[34]Gregory Fournier and Anthony Poole have proposed that Asgard is part of "the Eukaryote tree", forming a superphylum they call "Eukaryomorpha" defined by "shared derived characters" (eukaryote signature proteins).[35]
The taxonomy is uncertain and the phylum names are therefore somewhat speculative. The list of phyla is based on theList of Prokaryotic names with Standing in Nomenclature(LPSN)[36]andNational Center for Biotechnology Information(NCBI).[37]
- PhylumBaldrarchaeotaCaceres 2019
- PhylumBorrarchaeotaLiu et al. 2021
- PhylumFreyrarchaeotacorrig. Caceres 2019
- PhylumFriggarchaeotaCaceres 2019
- PhylumGefionarchaeotaCaceres 2019
- PhylumGerdarchaeotaCai et al. 2020
- PhylumHeimdallarchaeotaZaremba-Niedzwiedzka et al. 2017
- PhylumHelarchaeotaSeitz et al. 2019
- PhylumHermodarchaeotaLiu et al. 2021
- PhylumHodarchaeotaLiu et al. 2021
- PhylumIdunnarchaeotaCaceres 2019
- PhylumKariarchaeotaLiu et al. 2021
- PhylumLokiarchaeotaSpang et al. 2015
- PhylumNjordarchaeotaXie et al. 2022
- PhylumOdinarchaeotaZaremba-Niedzwiedzka et al. 2017
- PhylumSifarchaeotaFarag et al. 2020
- PhylumSigynarchaeotaXie et al. 2022
- PhylumThorarchaeotaBaker 2015
- PhylumTyrarchaeotaXie et al. 2022
- PhylumWukongarchaeotaLiu et al. 2021
Genomic elements
[edit]Viruses
[edit]Several family-level groups of viruses associated with Asgard archaea have been discovered using metagenomics.[38][39][40]The viruses were assigned to Lokiarchaeia, Thorarchaeia, Odinarchaeia and Helarchaeia hosts using CRISPR spacer matching to the corresponding protospacers within the viral genomes. Two groups of viruses (called 'verdandiviruses') are related to archaeal and bacterial viruses of the classCaudoviricetes,i.e., viruses with icosahedral capsids and helical tails;[38][40]two other distinct groups (called 'skuldviruses') are distantly related to tailless archaeal and bacterial viruses with icosahedral capsids of the realmVaridnaviria;[38][39]and the third group of viruses (calledwyrdviruses) is related to archaea-specific viruses with lemon-shaped virus particles (familyHalspiviridae).[38][39]The viruses have been identified in deep-sea sediments[38][40]and a terrestrial hot spring of the Yellowstone National Park.[39]All these viruses display very low sequence similarity to other known viruses but are generally related to the previously described prokaryotic viruses,[41]with no meaningful affinity to viruses of eukaryotes.[42][38]
Mobile genetic elements
[edit]In addition to viruses, several groups of crypticmobile genetic elementshave been discovered throughCRISPRspacer matching to be associated with Asgard archaea of the Lokiarchaeia, Thorarchaeia and Heimdallarchaeia lineages.[38][43]These mobile elements do not encode recognizable viral hallmark proteins and could represent either novel types of viruses or plasmids.
See also
[edit]References
[edit]- ^Fournier, G.P.; Poole, A.M. (2018)."A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree".Frontiers in Microbiology.9:1896.doi:10.3389/fmicb.2018.01896.PMC6104171.PMID30158917.
- ^Da Cunha, Violette; Gaia, Morgan; Gadelle, Daniele; et al. (June 2017)."Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes".PLOS Genetics.13(6): e1006810.doi:10.1371/journal.pgen.1006810.PMC5484517.PMID28604769.
- ^abcdZaremba-Niedzwiedzka, Katarzyna; Caceres, Eva F.; Saw, Jimmy H.; et al. (January 2017). "Asgard archaea illuminate the origin of eukaryotic cellular complexity".Nature.541(7637): 353–358.Bibcode:2017Natur.541..353Z.doi:10.1038/nature21031.OSTI1580084.PMID28077874.S2CID4458094.
- ^abEme, Laura; Spang, Anja; Lombard, Jonathan; Stairs, Courtney W.; Ettema, Thijs J. G. (November 2017). "Archaea and the origin of eukaryotes".Nature Reviews. Microbiology.15(12): 711–723.doi:10.1038/nrmicro.2017.133.PMID29123225.S2CID8666687.
- ^abWilliams, Tom A.; Cox, Cymon J.; Foster, Peter G.; Szöllősi, Gergely J.; Embley, T. Martin (January 2020)."Phylogenomics provides robust support for a two-domains tree of life".Nature Ecology & Evolution.4(1): 138–147.doi:10.1038/s41559-019-1040-x.PMC6942926.PMID31819234.
- ^Nobs, Stephanie-Jane; MacLeod, Fraser I.; Wong, Hon Lun; Burns, Brendan P. (May 2022). "Eukarya the chimera: eukaryotes, a secondary innovation of the two domains of life?".Trends in Microbiology.30(5): 421–431.doi:10.1016/j.tim.2021.11.003.PMID34863611.S2CID244823103.
- ^Doolittle, W. Ford (February 2020)."Evolution: Two Domains of Life or Three?".Current Biology.30(4): R177–R179.Bibcode:2020CBio...30.R177D.doi:10.1016/j.cub.2020.01.010.PMID32097647.
- ^Jørgensen, Steffen Leth; Hannisdal, Bjarte; Lanzen, Anders; et al. (October 2012)."Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge".PNAS.109(42): E2846–E2855.doi:10.1073/pnas.1207574109.PMC3479504.PMID23027979.
- ^Jørgensen, Steffen Leth; Thorseth, Ingunn H.; Pedersen, Rolf B.; et al. (October 4, 2013)."Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge".Frontiers in Microbiology.4:299.doi:10.3389/fmicb.2013.00299.PMC3790079.PMID24109477.
- ^abSpang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; et al. (May 2015)."Complex archaea that bridge the gap between prokaryotes and eukaryotes".Nature.521(7551): 173–179.Bibcode:2015Natur.521..173S.doi:10.1038/nature14447.PMC4444528.PMID25945739.
- ^Yong, Ed."Break in the Search for the Origin of Complex Life".The Atlantic.Retrieved2018-03-21.
- ^von Schnurbein, Stefanie (November 2000). "The Function of Loki in Snorri Sturluson's" Edda "".History of Religions.40(2): 109–124.doi:10.1086/463618.
- ^Spang, Anja; Eme, Laura; Saw, Jimmy H.; et al. (March 2018)."Asgard archaea are the closest prokaryotic relatives of eukaryotes".PLOS Genetics.14(3): e1007080.doi:10.1371/journal.pgen.1007080.PMC5875740.PMID29596421.
- ^Seitz, Kiley W.; Lazar, Cassandre S.; Hinrichs, Kai-Uwe; et al. (July 2016)."Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction".The ISME Journal.10(7): 1696–1705.Bibcode:2016ISMEJ..10.1696S.doi:10.1038/ismej.2015.233.PMC4918440.PMID26824177.
- ^Rodrigues-Oliveira, Thiago; Wollweber, Florian; Ponce-Toledo, Rafael I.; et al. (2023-01-12)."Actin cytoskeleton and complex cell architecture in an Asgard archaeon".Nature.613(7943): 332–339.Bibcode:2023Natur.613..332R.doi:10.1038/s41586-022-05550-y.ISSN0028-0836.PMC9834061.PMID36544020.
- ^Akıl, Caner; Robinson, Robert C. (October 2018). "Genomes of Asgard archaea encode profilins that regulate actin".Nature.562(7727): 439–443.Bibcode:2018Natur.562..439A.doi:10.1038/s41586-018-0548-6.PMID30283132.S2CID52917038.
- ^Akıl, Caner; Tran, Linh T.; Orhant-Prioux, Magali; Baskaran, Yohendran; Manser, Edward; Blanchoin, Laurent; Robinson, Robert C. (August 2020)."Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea".Proceedings of the National Academy of Sciences of the United States of America.117(33): 19904–19913.Bibcode:2020PNAS..11719904A.bioRxiv10.1101/768580.doi:10.1073/pnas.2009167117.PMC7444086.PMID32747565.
- ^Akıl, Caner; Ali, Samson; Tran, Linh T.; et al. (March 2022)."Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution".Science Advances.8(12): eabm2225.Bibcode:2022SciA....8M2225A.doi:10.1126/sciadv.abm2225.PMC8956254.PMID35333570.
- ^abcImachi, Hiroyuki; Nobu, Masaru K.; Nakahara, Nozomi; et al. (January 2020)."Isolation of an archaeon at the prokaryote-eukaryote interface".Nature.577(7791): 519–525.Bibcode:2020Natur.577..519I.doi:10.1038/s41586-019-1916-6.PMC7015854.PMID31942073.
- ^Penev, Petar I.; Fakhretaha-Aval, Sara; Patel, Vaishnavi J.; et al. (October 2020)."Supersized Ribosomal RNA Expansion Segments in Asgard Archaea".Genome Biology and Evolution.12(10): 1694–1710.doi:10.1093/gbe/evaa170.PMC7594248.PMID32785681.
- ^Tirumalai, Madhan R.; Raghavan, Sivaraman V.; Kutty, Layla A.; Song, Eric L.; Fox, George E. (October 2023)."Ribosomal Protein Cluster Organization in Asgard Archaea".Archaea.2023:16.doi:10.1155/2023/5512414.PMC10833476.PMID38314098.
- ^abcdMacLeod, Fraser; Kindler, Gareth S.; Wong, Hon Lun; Chen, Ray; Burns, Brendan P. (2019)."Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes".AIMS Microbiology.5(1): 48–61.doi:10.3934/microbiol.2019.1.48.PMC6646929.PMID31384702.
- ^abLiu, Yang; Makarova, Kira S.; Huang, Wen-Cong; et al. (2020)."Expanding diversity of Asgard archaea and the elusive ancestry of eukaryotes".bioRxiv.doi:10.1101/2020.10.19.343400.S2CID225056970.
- ^Cai, Mingwei; Richter-Heitmann, Tim; Yin, Xiuran; et al. (2021). "Ecological features and global distribution of Asgard archaea".Science of the Total Environment.758:143581.Bibcode:2021ScTEn.75843581C.doi:10.1016/j.scitotenv.2020.143581.ISSN0048-9697.PMID33223169.S2CID227134171.
- ^Henneman, Bram; van Emmerik, Clara; van Ingen, Hugo; Dame, Remus T. (September 2018)."Structure and function of archaeal histones".PLOS Genetics.14(9): e1007582.Bibcode:2018BpJ...114..446H.doi:10.1371/journal.pgen.1007582.PMC6136690.PMID30212449.
- ^Zimmer, Carl(15 January 2020)."This Strange Microbe May Mark One of Life's Great Leaps - A organism living in ocean muck offers clues to the origins of the complex cells of all animals and plants".The New York Times.Retrieved16 January2020.
- ^Liu, Yang; Makarova, Kira S.; Huang, Wen-Cong; Wolf, Yuri I.; Nikolskaya, Anastasia; Zhang, Xinxu; et al. (May 2021)."Expanded diversity of Asgard archaea and their relationships with eukaryotes".Nature.593(7860): 553–557.Bibcode:2021Natur.593..553L.doi:10.1038/s41586-021-03494-3.PMC11165668.PMID33911286.S2CID233447651.
- ^"GTDB release 08-RS214".Genome Taxonomy Database.Retrieved10 May2023.
- ^"ar53_r214.sp_label".Genome Taxonomy Database.Retrieved10 May2023.
- ^"Taxon History".Genome Taxonomy Database.Retrieved10 May2023.
- ^Eme, Laura; Tamarit, Daniel; Caceres, Eva F.; et al. (2023-06-14)."Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes".Nature.618(7967): 992–999.Bibcode:2023Natur.618..992E.doi:10.1038/s41586-023-06186-2.PMC10307638.PMID37316666.
- ^Latorre, A.; Durban, A.; Moya, A.; Pereto, J. (2011)."The role of symbiosis in eukaryotic evolution".In Gargaud, M.; López-Garcìa, P.; Martin H. (eds.).Origins and Evolution of Life: An astrobiological perspective.Cambridge:Cambridge University Press.pp. 326–339.ISBN978-0-521-76131-4.Archivedfrom the original on 24 March 2019.Retrieved27 August2017.
- ^López-García, Purificación; Moreira, David (July 2019)."Eukaryogenesis, a syntrophy affair".Nature Microbiology.4(7): 1068–1070.doi:10.1038/s41564-019-0495-5.PMC6684364.PMID31222170.
- ^Rodrigues-Oliveira, Thiago; Wollweber, Florian; Ponce-Toledo, Rafael I.; Xu, Jingwei; Rittmann, Simon K.-M. R.; Klingl, Andreas; Pilhofer, Martin; Schleper, Christa (January 2023)."Actin cytoskeleton and complex cell architecture in an Asgard archaeon".Nature.613(7943): 332–339.Bibcode:2023Natur.613..332R.doi:10.1038/s41586-022-05550-y.ISSN1476-4687.PMC9834061.PMID36544020.
- ^Fournier, Gregory P.; Poole, Anthony M. (2018-08-15)."A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree".Frontiers in Microbiology.9:1896.doi:10.3389/fmicb.2018.01896.ISSN1664-302X.PMC6104171.PMID30158917.
- ^Euzéby, J.P."Superphylum" Asgardarchaeota "".List of Prokaryotic names with Standing in Nomenclature(LPSN).Retrieved2021-06-27.
- ^"Asgard group".National Center for Biotechnology Information(NCBI) taxonomy database.Retrieved2021-03-20.
- ^abcdefgMedvedeva, S.; Sun, J.; Yutin, N.;Koonin, Eugene V.;Nunoura, T.; Rinke, C.; Krupovic, M. (July 2022)."Three families of Asgard archaeal viruses identified in metagenome-assembled genomes".Nature Microbiology.7(7): 962–973.doi:10.1038/s41564-022-01144-6.PMC11165672.PMID35760839.S2CID250091635.
- ^abcdTamarit, D.; Caceres, E.F.; Krupovic, M.; Nijland, R.; Eme, L.; Robinson, N.P.; Ettema, T.J.G. (July 2022)."A closed Candidatus Odinarchaeum chromosome exposes Asgard archaeal viruses".Nature Microbiology.7(7): 948–952.doi:10.1038/s41564-022-01122-y.PMC9246712.PMID35760836.S2CID250090798.
- ^abcRambo, I.M.; Langwig, M.V.; Leão, P.; De Anda, V.; Baker, B.J. (July 2022)."Genomes of six viruses that infect Asgard archaea from deep-sea sediments".Nature Microbiology.7(7): 953–961.doi:10.1038/s41564-022-01150-8.PMID35760837.
- ^Prangishvili, D.; Bamford, D.H.; Forterre, P.; Iranzo, J.; Koonin, Eugene V.; Krupovic, M. (November 2017). "The enigmatic archaeal virosphere".Nature Reviews. Microbiology.15(12): 724–739.doi:10.1038/nrmicro.2017.125.PMID29123227.S2CID21789564.
- ^Alarcón-Schumacher, T.; Erdmann, S. (July 2022). "A trove of Asgard archaeal viruses".Nature Microbiology.7(7): 931–932.doi:10.1038/s41564-022-01148-2.PMID35760838.S2CID250091028.
- ^Wu, F.; Speth, D.R.; Philosof, A.; et al. (February 2022)."Unique mobile elements and scalable gene flow at the prokaryote-eukaryote boundary revealed by circularized Asgard archaea genomes".Nature Microbiology.7(2): 200–212.doi:10.1038/s41564-021-01039-y.PMC8813620.PMID35027677.
External links
[edit]- Traci Watson:The trickster microbes that are shaking up the tree of life,in:Nature,14 May 2019

![Metabolic pathways of Asgard archaea, varying by phyla[22]](https://upload.wikimedia.org/wikipedia/commons/thumb/7/7f/Asgard_archaea_Phyla_%28cropped%29.png/634px-Asgard_archaea_Phyla_%28cropped%29.png)
![Metabolic pathways of Asgard archaea, varying by environment[22]](https://upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Asgard_archaea_in_various_environments_%28cropped%29.png/638px-Asgard_archaea_in_various_environments_%28cropped%29.png)

