Lokiarchaeota
This article needs to beupdated.(March 2018) |
| Lokiarchaeota | |
|---|---|
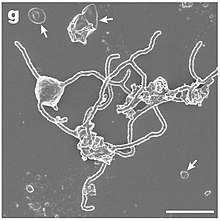
| |
| SEM image ofCandidatusPrometheoarchaeum syntrophicum | |
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Superphylum: | |
| Phylum: | "Lokiarchaeota" Spang et al. 2015
|
| Class: | "Lokiarchaeia" corrig. Spang et al. 2015
|
| Order | |
| |
Lokiarchaeotais a proposedphylumof theArchaea.[1]The phylum includes all members of the group previously named Deep Sea Archaeal Group, also known as Marine Benthic Group B. Lokiarchaeota is part of the superphylumAsgardcontaining the phyla: Lokiarchaeota,Thorarchaeota,Odinarchaeota,Heimdallarchaeota,andHelarchaeota.[2]Aphylogeneticanalysis disclosed amonophyleticgrouping of the Lokiarchaeota with theeukaryotes.The analysis revealed severalgeneswithcell membrane-related functions. The presence of such genes support the hypothesis of anarchaealhost for the emergence of the eukaryotes; theeocyte-like scenarios.
Lokiarchaeota was introduced in 2015 after the identification of a candidategenomein ametagenomicanalysis of a mid-oceanic sediment sample. This analysis suggests the existence of agenusof unicellular life dubbedLokiarchaeum.The sample was taken near ahydrothermal ventat a vent field known asLoki's Castlelocated at the bend betweenMohns/Knipovich ridge in theArctic Ocean.[3]
Discovery
[edit]Sediments from a gravitycoretaken in 2010 in the rift valley on the Knipovich ridge in the Arctic Ocean, near the so-calledLoki's Castlehydrothermal ventsite, were analysed. Specific sediment horizons, previously shown to contain high abundances of novel archaeal lineages[4][5]were subjected to metagenomic analysis. Due to the low density of cells in the sediment, the resulting genetic sequence does not come from an isolated cell, as would be the case in conventional analysis, but is rather a combination of genetic fragments.[6]The result was a 92% complete, 1.4 fold-redundant composite genome namedLokiarchaeum.[3]
The metagenomic analysis determined the presence of an organism's genome in the sample.[3]However, the organism itself was not cultured until years later, with a Japanese group first reporting isolation and cultivation of a Lokiarchaeota strain in 2019.[7]Since this initial cultivation of Lokiarchaeota, members of the phylum have been reported in a diverse range of habitats. Advances in both long and short-read technologies for DNA sequencing have also aided in the recovery and identification of Lokiarchaeota from microbial samples.[2]
The Lokiarchaeota phylum was proposed based onphylogeneticanalyses using a set of highly conserved protein-coding genes.[3]Through a reference to the hydrothermal vent complex from which the first genome sample originated, the name refers toLoki,the Norse shape-shifting god.[6]The Loki of literature has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies",[8]an analogy to the role of Lokiarchaeota in debates about the origin of eukaryotes.[3]
Description
[edit]
TheLokiarchaeumcomposite genome consists of 5,381 protein codinggenes.Of these, roughly 32% do not correspond to any known protein, 26% closely resemble archaeal proteins, and 29% correspond to bacterial proteins. This situation is consistent with: (i) proteins from a novel phylum (with few close relatives, or none) being difficult to assign to their correctdomain;and (ii) existing research that suggests there has been significant inter-domaingene transferbetweenbacteriaand Archaea.
A small, but significant portion of the proteins (175, 3.3%) that the recovered genes code for are very similar toeukaryoticproteins. These proteins included homologs of cytoskeleton proteins, GTPases, and the oligosaccharyltransferase (OST) protein complex. Homologues for components of the endosomal sorting complex required for transport and the ubiquitin protein modifier system were also identified in Lokiarchaeota genome analysis.[2]Sample contamination is an unlikely explanation for the unusual proteins because the recovered genes were always flanked byprokaryoticgenes and no genes of known eukaryotic origin were detected in the metagenome from which the composite genome was extracted. Further, previousphylogeneticanalysis suggested the genes in question had their origin at the base of the eukaryoticclades.[3]
In eukaryotes, the function of these shared proteins includecell membranedeformation, cell shape formation, and a dynamic proteincytoskeleton.[3][9][10]Eukaryotic protein functions found in Lokiarchaeota also include intracellular transport mechanisms.[11]It is inferred then thatLokiarchaeummay have some of these abilities.[3]Another shared protein,actin,is essential forphagocytosisin eukaryotes.[6][9]Phagocytosis is the ability to engulf and consume another particle; such ability would facilitate theendosymbiotic originofmitochondriaandchloroplasts,which is a key difference between prokaryotes and eukaryotes.[3]The presence of actin proteins and intracellular transport mechanisms provides evidence for the common ancestry between ancient Lokiarchaeota and eukarya.[11]
Taxonomy
[edit]- Order"Lokiarchaeales"Spang et al. 2015[12][13]
- Family"Lokiarchaeaceae"Vanwonterghem et al. 2016(MBGB, DSAG)
- Genus "CandidatusLokiarchaeum"corrig. Spang et al. 2015
- "Ca.L. ossiferum "Rodrigues-Oliveira et al. 2023
- Genus "CandidatusLokiarchaeum"corrig. Spang et al. 2015
- Family"Prometheoarchaeaceae"Pallen, Rodriguez-R & Alikhan 2022(MK-D1)
- Genus "CandidatusHarpocratesius"Wu et al. 2022
- "Ca.H. repetitus "Wu et al. 2022
- Genus "CandidatusPromethearchaeum"corrig. Imachi, Nobu & Takai 2020
- "Ca.P. syntrophicum "corrig. Imachi, Nobu & Takai 2020
- Genus "CandidatusHarpocratesius"Wu et al. 2022
- Family"Lokiarchaeaceae"Vanwonterghem et al. 2016(MBGB, DSAG)
Evolutionary significance
[edit]
A comparative analysis of theLokiarchaeumgenome against known genomes resulted in aphylogenetic treethat showed amonophyleticgroup composed of the Lokiarchaeota and the eukaryotes,[14]supporting an archaeal host oreocyte-like scenariosfor the emergence of the eukaryotes.[15][16][17]The repertoire of membrane-related functions ofLokiarchaeumsuggests that thecommon ancestorto the eukaryotes might be an intermediate step between the prokaryotic cells, devoid of subcellular structures, and the eukaryotic cells, which harbor manyorganelles.[3]
Carl Woese'sthree-domain systemclassifies cellular life into three domains: archaea, bacteria, and eukaryotes; the last being characterised by large, highly evolved cells, containingmitochondria,which help the cells produceATP(adenosine triphosphate, the energy currency of the cell), and a membrane-boundnucleuscontainingnucleic acids.Protozoaand all multicellular organisms such as animals, fungi, and plants are eukaryotes.
The bacteria and archaea are thought to be the most ancient of lineages,[18]as fossil strata bearing the chemical signature of archaeallipidshave been dated back to 3.8 billion years ago.[19]The eukaryotes include all complex cells and almost all multicellular organisms. They are thought to have evolved between 1.6 and 2.1 billion years ago.[20]While the evolution of eukaryotes is considered to be an event of great evolutionary significance, no intermediate forms or "missing links" had been discovered previously. In this context, the discovery ofLokiarchaeum,with some but not all of the characteristics of eukaryotes, provides evidence on the transition from archaea to eukaryotes.[21]Lokiarchaeota and the eukaryotes probably share a common ancestor, and if so, diverged roughly two billion years ago. Evidence for common ancestry, rather than an evolutionary shift from Lokiarchaeota to eukaryotes, is found in analysis of fold superfamilies (FSFs). Fold super families are evolutionarily defined domains of protein structure. It is estimated that there are around 2500 total FSFs found in nature.[11]Utilization of Venn diagrams allowed researchers to depict distributions of FSFs of those that were shared by Archaea and Eukarya, as well as those unique to their respective kingdoms. The addition of Lokiarchaeum into the Venn groups created from an initial genomic census only added 10 FSFs to Archaea. The addition of Lokiarchaeum also only contributed to a decrease of two FSFs previously unique to Eukarya. There were still 284 FSFs found exclusively in Eukarya. Lokiarchaeota’s limited impact in changing the Venn distribution of FSFs demonstrates the lack of genes that could be traced to a common ancestor with Eukaryotes. Rather, Eukaryotic genes present in bacterial and archaeal organisms are hypothesized to be from horizontal transfer from an early ancestor of modern eukaryotes.[11]This putative ancestor possessed crucial "starter" genes that enabled increased cellular complexity. This common ancestor, or a relative, eventually led to the evolution of eukaryotes.[6]
In 2020, a Japanese research group reported culturing a strain of Lokiarchaeota in the laboratory.[22][23][7]This strain, currently namedCandidatus Prometheoarchaeum syntrophicumstrain MK-D1, was observed insyntrophicassociation with two hydrogen-consuming microbes: asulfate-reducing bacteriaof the genusHalodesulfovibrioand amethanogenof the genusMethanogenium.The MK-D1 organism produces hydrogen as a metabolic byproduct, which is then consumed by thesymbioticsyntrophs. MK-D1 also seems to organize its external membrane into complex structures using genes shared with eukaryotes. While association withalphaproteobacteria(from which mitochondria are thought to descend) was not observed, these features suggest that MK-D1 and its syntrophs may represent an extant example of archaea-bacteria symbiosis similar to that which gave rise to eukaryotes.
In 2022, the second cultured example of Lokiarchaeota was reported and the strain was namedCandidatus Lokiarchaeum ossiferum.[24]
Physiological traits
[edit]Lokiarachaeota is known to have a tetrahydromethanopterin-dependentWood-Ljundahlpathway. This pathway contains a series of biochemical reactions aiding in inorganic carbon utilization. In Lokiarchaeota, the WLP is thought to be acetogenic, due to lacking the gene methyl-CoM reductase necessary for methanogenesis.[2]
Analysis of Lokiarchaeon genes also showed the expression of protein-encoding open reading frames (ORFs) involving the metabolism of sugars and proteins. However, these metabolic activities vary between subgroups of Lokiarchaeota. While Lokiarchaeota subgroups have similar genetic information, differences in metabolic abilities explain their respective ecological niches.[25]
Two major subgroups of the Lokiarachaeota phylum are Loki-2 and Loki-3. Incubations of these two subgroups from Helgoland mud sediments were analyzed through RNA and DNA stable isotope probing to understand their respective carbon metabolisms.[25]Loki-3 were found to be active in both organic carbon utilization and the degradation of aromatic compounds. The Loki-3 subgroup was not found to utilize proteins or short chain fatty acids, even though genes for amino acid degradation were present in both subgroups. Loki-2 was found to utilize protein, as seen through activity in when proteins were provided in Loki-2 incubations. Due to the greater carbon utilization pathways of Loki-3, the subgroup is found in a more diverse range of marine sediments than Loki-2.[25]
See also
[edit]References
[edit]- ^Lambert, Jonathan (9 August 2019)."Scientists glimpse oddball microbe that could help explain rise of complex life - 'Lokiarchaea', previously known only from DNA, is isolated and grown in culture".Nature.572(7769): 294.Bibcode:2019Natur.572..294L.doi:10.1038/d41586-019-02430-w.PMID31409927.
- ^abcdCaceres, Eva F.; Lewis, William H.; Homa, Felix; Martin, Tom; Schramm, Andreas; Kjeldsen, Kasper U.; Ettema, Thijs J. G. (2019-12-18)."Near-complete Lokiarchaeota genomes from complex environmental samples using long and short read metagenomic analyses":2019.12.17.879148.doi:10.1101/2019.12.17.879148.S2CID213982145.
{{cite journal}}:Cite journal requires|journal=(help) - ^abcdefghijkSpang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; van Eijk, Roel; Schleper, Christa; Guy, Lionel; Ettema, Thijs J. G. (2015)."Complex archaea that bridge the gap between prokaryotes and eukaryotes".Nature.521(7551): 173–179.Bibcode:2015Natur.521..173S.doi:10.1038/nature14447.ISSN0028-0836.PMC4444528.PMID25945739.
- ^Jørgensen, Steffen Leth; Hannisdal, Bjarte; Lanzen, Anders; Baumberger, Tamara; Flesland, Kristin; Fonseca, Rita; Øvreås, Lise; Steen, Ida H; Thorseth, Ingunn H; Pedersen, Rolf B; Schleper, Christa (September 5, 2012)."Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge".PNAS.109(42): E2846–55.doi:10.1073/pnas.1207574109.PMC3479504.PMID23027979.
- ^Jørgensen, Steffen Leth; Thorseth, Ingunn H; Pedersen, Rolf B; Baumberger, Tamara; Schleper, Christa (October 4, 2013)."Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge".Frontiers in Microbiology.4:299.doi:10.3389/fmicb.2013.00299.PMC3790079.PMID24109477.
- ^abcdRincon, Paul (May 6, 2015)."Newly found microbe is close relative of complex life".BBC.RetrievedMay 9,2015.
- ^abImachi, Hiroyuki; et al. (15 January 2020)."Isolation of an archaeon at the prokaryote–eukaryote interface".Nature.577(7791): 519–525.Bibcode:2020Natur.577..519I.bioRxiv10.1101/726976.doi:10.1038/s41586-019-1916-6.PMC7015854.PMID31942073.
- ^von Schnurbein, Stefanie (November 2000). "The Function of Loki in Snorri Sturluson's" Edda "".History of Religions.40(2): 109–124.doi:10.1086/463618.
- ^abGhoshdastider U, Jiang S, Popp D, Robinson RC (2015)."In search of the primordial actin filament".Proc Natl Acad Sci U S A.112(30): 9150–1.doi:10.1073/pnas.1511568112.PMC4522752.PMID26178194.
- ^Khan, Amina (May 6, 2015)."Meet Loki, your closest-known prokaryote relative".LA Times.RetrievedMay 9,2015.
- ^abcdNasir, Arshan; Kim, Kyung Mo; Caetano-Anollés, Gustavo (2015-08-01)."Lokiarchaeota: eukaryote-like missing links from microbial dark matter?".Trends in Microbiology.23(8): 448–450.doi:10.1016/j.tim.2015.06.001.ISSN0966-842X.PMID26112912.
- ^J.P. Euzéby."Family: Lokiarchaeota, not assigned to family".List of Prokaryotic names with Standing in Nomenclature(LPSN).Retrieved2023-06-20.
- ^Sayers; et al."Candidatus Lokiarchaeota".National Center for Biotechnology Information(NCBI) taxonomy database.Retrieved2023-06-20.
- ^Maximillan, Ludwig (27 December 2019)."Evolution: A revelatory relationship".Phys.org.Retrieved28 December2019.
- ^Embley, T. Martin;Martin, William (2006)."Eukaryotic evolution, changes and challenges".Nature.440(7084): 623–630.Bibcode:2006Natur.440..623E.doi:10.1038/nature04546.ISSN0028-0836.PMID16572163.S2CID4396543.
- ^Lake, James A. (1988). "Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences".Nature.331(6152): 184–186.Bibcode:1988Natur.331..184L.doi:10.1038/331184a0.ISSN0028-0836.PMID3340165.S2CID4368082.
- ^Guy, Lionel; Ettema, Thijs J.G. (2011). "The archaeal 'TACK' superphylum and the origin of eukaryotes".Trends in Microbiology.19(12): 580–587.doi:10.1016/j.tim.2011.09.002.ISSN0966-842X.PMID22018741.
- ^Wang, Minglei; Yafremava, Liudmila S.; Caetano-Anollés, Derek; Mittenthal, Jay E.; Caetano-Anollés, Gustavo (2007)."Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world".Genome Research.17(11): 1572–1585.doi:10.1101/gr.6454307.PMC2045140.PMID17908824.
- ^Hahn, Jürgen; Haug, Pat (1986). "Traces of Archaebacteria in ancient sediments".Systematic and Applied Microbiology.7(Archaebacteria '85 Proceedings): 178–83.doi:10.1016/S0723-2020(86)80002-9.
- ^Knoll, Andrew H.; Javaux, E. J.; Hewitt, D.; Cohen, P. (29 June 2006)."Eukaryotic organisms in Proterozoic oceans".Philosophical Transactions of the Royal Society B.361(1470): 1023–38.doi:10.1098/rstb.2006.1843.PMC1578724.PMID16754612.
- ^Zimmer, Carl (6 May 2015)."Under the Sea, a Missing Link in the Evolution of Complex Cells".New York Times.Retrieved8 May2015.
- ^Timmer, John (2019-08-07)."We've finally gotten a look at the microbe that might have been our ancestor".Ars Technica.Retrieved2019-08-08.
- ^Zimmer, Carl (2020-01-15)."This Strange Microbe May Mark One of Life's Great Leaps".The New York Times.Retrieved2020-01-15.
- ^Rodrigues-Oliveira, Thiago; Wollweber, Florian; Ponce-Toledo, Rafael I.; Xu, Jingwei; Rittmann, Simon K.-M. R.; Klingl, Andreas; Pilhofer, Martin; Schleper, Christa (January 2023)."Actin cytoskeleton and complex cell architecture in an Asgard archaeon".Nature.613(7943): 332–339.doi:10.1038/s41586-022-05550-y.ISSN1476-4687.PMC9834061.PMID36544020.
- ^abcYin, Xiuran; Cai, Mingwei; Liu, Yang; Zhou, Guowei; Richter-Heitmann, Tim; Aromokeye, David A.; Kulkarni, Ajinkya C.; Nimzyk, Rolf; Cullhed, Henrik; Zhou, Zhichao; Pan, Jie (Mar 2021)."Subgroup level differences of physiological activities in marine Lokiarchaeota".The ISME Journal.15(3): 848–861.doi:10.1038/s41396-020-00818-5.ISSN1751-7370.PMC8027215.PMID33149207.
External links
[edit]- Sokol, Joshua (2023-04-11)."Primitive Asgard Cells Show Life on the Brink of Complexity".Quanta Magazine.Retrieved2023-04-16.
