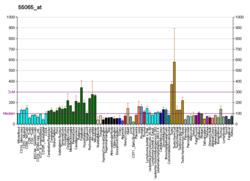
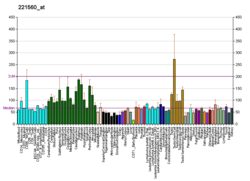
MARK4
MAP/microtubule affinity-regulating kinase 4is anenzymethat in humans is encoded by theMARK4gene.[5][6][7]MARK4 belongs to the family ofserine/threonine kinasesthat phosphorylatemicrotubule-associated proteins(MAP) causing their detachment frommicrotubules.[8]Detachment thereby increases microtubule dynamics and facilitates a number of cell activities includingcell division,cell cyclecontrol,cell polaritydetermination, and cell shape alterations.[9]
There are four members of the MARK protein family, MARK1-4, which are highly conserved. MARK4 kinase has been shown to be involved in microtubule organization in neuronal cells. Levels of MARK4 are elevated inAlzheimer's diseaseand may contribute to the pathological phosphorylation oftau proteinin this disease.
Interactions
[edit]MARK4 has been shown tointeractwithUSP9X[10]andUbiquitin C.[10]
References
[edit]- ^abcGRCh38: Ensembl release 89: ENSG00000007047–Ensembl,May 2017
- ^abcGRCm38: Ensembl release 89: ENSMUSG00000030397–Ensembl,May 2017
- ^"Human PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^"Mouse PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^Kato T, Satoh S, Okabe H, Kitahara O, Ono K, Kihara C, Tanaka T, Tsunoda T, Yamaoka Y, Nakamura Y, Furukawa Y (Apr 2001)."Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis".Neoplasia.3(1): 4–9.doi:10.1038/sj.neo.7900132.PMC1505019.PMID11326310.
- ^Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (April 1997)."MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption".Cell.89(2): 297–308.doi:10.1016/S0092-8674(00)80208-1.PMID9108484.
- ^"Entrez Gene: MARK4 MAP/microtubule affinity-regulating kinase 4".
- ^Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (April 1997)."MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption".Cell.89(2): 297–308.doi:10.1016/s0092-8674(00)80208-1.PMID9108484.
- ^Naz F, Anjum F, Islam A, Ahmad F, Hassan MI (November 2013). "Microtubule affinity-regulating kinase 4: structure, function, and regulation".Cell Biochemistry and Biophysics.67(2): 485–99.doi:10.1007/s12013-013-9550-7.PMID23471664.S2CID13507976.
- ^abAl-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR (April 2008)."Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains"(PDF).The Biochemical Journal.411(2): 249–60.doi:10.1042/BJ20080067.PMID18254724.S2CID13038944.
Further reading
[edit]- Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O (April 2001)."Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro".DNA Research.8(2): 85–95.doi:10.1093/dnares/8.2.85.PMID11347906.
- Beghini A, Magnani I, Roversi G, Piepoli T, Di Terlizzi S, Moroni RF, Pollo B, Fuhrman Conti AM, Cowell JK, Finocchiaro G, Larizza L (May 2003)."The neural progenitor-restricted isoform of the MARK4 gene in 19q13.2 is upregulated in human gliomas and overexpressed in a subset of glioblastoma cell lines".Oncogene.22(17): 2581–91.doi:10.1038/sj.onc.1206336.PMID12735302.
- Trinczek B, Brajenovic M, Ebneth A, Drewes G (February 2004)."MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes".The Journal of Biological Chemistry.279(7): 5915–23.doi:10.1074/jbc.M304528200.PMID14594945.
- Brajenovic M, Joberty G, Küster B, Bouwmeester T, Drewes G (March 2004)."Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network".The Journal of Biological Chemistry.279(13): 12804–11.doi:10.1074/jbc.M312171200.PMID14676191.
- Schneider A, Laage R, von Ahsen O, Fischer A, Rossner M, Scheek S, Grünewald S, Kuner R, Weber D, Krüger C, Klaussner B, Götz B, Hiemisch H, Newrzella D, Martin-Villalba A, Bach A, Schwaninger M (March 2004)."Identification of regulated genes during permanent focal cerebral ischaemia: characterization of the protein kinase 9b5/MARKL1/MARK4".Journal of Neurochemistry.88(5): 1114–26.doi:10.1046/j.1471-4159.2003.02228.x.PMID15009667.S2CID44856849.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J,Zoghbi HY,Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network".Nature.437(7062): 1173–8.Bibcode:2005Natur.437.1173R.doi:10.1038/nature04209.PMID16189514.S2CID4427026.
- Wissing J, Jänsch L, Nimtz M, Dieterich G, Hornberger R, Kéri G, Wehland J, Daub H (March 2007)."Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry".Molecular & Cellular Proteomics.6(3): 537–47.doi:10.1074/mcp.T600062-MCP200.hdl:10033/19756.PMID17192257.