ORF1ab
| Replicase polyprotein | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | rep | ||||||
| UniProt | P0C6X7 | ||||||
| |||||||
| Replicase polyprotein | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | rep | ||||||
| UniProt | P0DTD1 | ||||||
| |||||||
ORF1ab(alsoORF1a/b) refers collectively to twoopen reading frames(ORFs),ORF1aandORF1b,that are conserved in thegenomesofnidoviruses,a group of viruses that includescoronaviruses.Thegenesexpress largepolyproteinsthat undergoproteolysisto form severalnonstructural proteinswith various functions in theviral life cycle,includingproteasesand the components of thereplicase-transcriptase complex(RTC).[1][2][3]Together the two ORFs are sometimes referred to as thereplicase gene.[4]They are related by aprogrammed ribosomal frameshiftthat allows theribosometo continuetranslatingpast thestop codonat the end of ORF1a, in a -1reading frame.The resulting polyproteins are known aspp1aandpp1ab.[1][2][3][4]
Expression
[edit]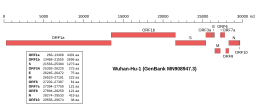 Genomicorganisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2, indicating the location of ORF1a and ORF1b | |
| NCBIgenome ID | 86693 |
|---|---|
| Genome size | 29,903 bases |
| Year of completion | 2020 |
| Genome browser(UCSC) | |
ORF1a is the firstopen reading frameat the5' endof the genome. Together ORF1ab occupies about two thirds of the genome, with the remaining third at the3' endencoding thestructural proteinsandaccessory proteins.[1][2][3]It is translated from a5' cappedRNA bycap-dependent translation.[1]Nidoviruses have a complex system of discontinuoussubgenomic RNAproduction to enable expression of genes in their relatively large RNA genomes (typically 27-32kbfor coronaviruses[1]), but ORF1ab is translated directly from the genomic RNA.[5]ORF1ab sequences have been observed in noncanonical subgenomic RNAs, though their functional significance is unclear.[5]
Aprogrammed ribosomal frameshiftallows reading through thestop codonthat terminates ORF1a to continue in a -1reading frame,producing the longer polyprotein pp1ab. The frameshift occurs at aslippery sequencewhich is followed by apseudoknotRNA secondary structure.[1]This has been measured at between 20-50% efficiency formurine coronavirus,[6]or 45-70% inSARS-CoV-2[7]yielding astoichiometryof roughly 1.5 to 2 times as much pp1a as pp1ab protein expressed.[2]
Processing
[edit]
Thepolyproteinspp1a and pp1ab contain about 13 to 17nonstructural proteins.[3]They undergo auto-proteolysisto release the nonstructural proteins due to the actions of internalcysteine proteasedomains.[1][2][3]
In coronaviruses, there are a total of 16 nonstructural proteins; pp1a protein containsnonstructural proteinsnsp1-11 and the pp1ab protein contains nsp1-10 and nsp12-16. Proteolytic processing is performed by two proteases: thepapain-like proteaseprotein domainlocated in the multidomain protein nsp3 cleaves up to nsp4, and the3CL protease(also known as the main protease, nsp5) performs the remaining cleavages of nsp5 through the polyproteinC-terminus.[1][2]Proteins nsp12-16, the C-terminal components of the pp1ab polyprotein, contain the coreenzymaticactivities necessary forviral replication.[1]After proteolytic processing, several of the nonstructural proteins assemble into a largeprotein complexknown as thereplicase-transcriptase complex(RTC) which performs genome replication andtranscription.[1][2]
Components
[edit]Core replicase domains
[edit]
A set of fiveconserved"core replicase"protein domainsare present in all nidovirus lineages (arteriviruses,mesoniviruses,roniviruses,andcoronaviruses): from ORF1a, themain proteaseflanked on either end bytransmembrane domains;and from ORF1b, anucleotidyltransferasedomain known asNiRAN,RNA-dependent RNA polymerase(RdRp), azinc-binding domain, and ahelicase.[3][9](This is sometimes considered seven domains, counting the transmembrane regions separately.[4]) In addition, anendoribonucleasedomain is found in all nidoviruses that infectvertebratehosts. Arteriviruses, which have smaller genomes than the other nidovirus lineages, also lackmethyltransferasesas well as a proofreadingexoribonuclease,a domain that is conserved in nidoviruses with larger genomes.[3]This proofreading functionality is thought to be required for sufficient fidelity to replicate large RNA genomes, but may also play additional roles in some viruses.[9]
Coronaviruses
[edit]In coronaviruses, pp1a and pp1ab together contain sixteen nonstructural proteins, which have the following functions:[1][2][10][11]
| Nonstructural protein | Function |
|---|---|
| nonstructural protein 1 | CellularmRNAdegradation,host celltranslation inhibition,interferoninhibition; not present inGammacoronavirus |
| nonstructural protein 2 | Unknown; bindsprohibitin |
| nonstructural protein 3 | Multi-domainprotein with one or twopapain-like proteasedomains for polyprotein processing; interferon antagonist; multiple other roles |
| nonstructural protein 4 | Double-membranevesicleformation |
| nonstructural protein 5 | 3CL proteasefor polyprotein processing; interferon inhibition |
| nonstructural protein 6 | Double-membranevesicleformation |
| nonstructural protein 7 | Cofactor andprocessivityfactor forRdRp;forms complex with nsp8 and nsp12 |
| nonstructural protein 8 | Cofactor andprocessivityfactor forRdRp;forms complex with nsp7 and nsp12 |
| nonstructural protein 9 | Single-stranded RNA binding |
| nonstructural protein 10 | Cofactor for nsp14 and nsp16 |
| nonstructural protein 11 | Unknown |
| nonstructural protein 12 | RNA-dependent RNA polymerase(RdRp) andnucleotidyltransferase |
| nonstructural protein 13 | HelicaseandRNA triphosphatase |
| nonstructural protein 14 | Proofreadingexonuclease,RNA cap formation,guanosineN7-methyltransferase |
| nonstructural protein 15 | Endoribonuclease,immune evasionfunction |
| nonstructural protein 16 | Ribose2'-O-methyltransferase,RNA cap formation |
Evolution
[edit]The structure and organization of the genome, including ORF1a, ORF1b, and theframeshiftseparating them, is conserved among nidoviruses. Some "non-canonical" nidovirus structures have been described, mainly involvinggene fusions.[4]The largest known nidovirus,planarian secretory cell nidovirus(PSCNV), with a 41kb genome, has a non-canonical genome structure in which ORF1a, ORF1b, and downstream ORFs containing structural proteins are fused and expressed as a single large ORF encoding a polyprotein of over 13,000amino acids.[4][12]In these non-canonical genomes, other frameshift locations orstop codonreadthrough may be used to regulate thestoichiometryof viral proteins.[4]
Nidoviruses vary widely in genome size, fromarteriviruseswith typically 12-15kb genomes tocoronavirusesat 27-32kb. Their evolutionary history has been of research interest in understanding the replication of very large RNA genomes despite the relatively low-fidelity replication mechanism of the viralRNA-dependent RNA polymerase(RdRp).[4]The larger nidovirus genomes (above around 20kb[3]) encode a proofreadingexoribonuclease(nsp14in coronaviruses) thought to be required for replication fidelity.[9][1]
Amongcoronaviruses,ORF1ab is more highly conserved than the 3' ORFs encodingstructural proteins.[11]Throughout theCOVID-19 pandemic,thegenomeofSARS-CoV-2viruses has beensequencedmany times, resulting in identification of thousands of distinctvariants.In aWorld Health Organizationanalysis from July 2020, ORF1ab was the most frequentlymutatedgene, followed by the S gene encoding thespike protein.The most commonly mutated protein within ORF1ab waspapain-like protease(nsp3), and the single most commonly observedmissense mutationwas inRNA-dependent RNA polymerase.[13]SomePCRtests that detect COVID-19 analyze the specimen for the ORF1ab gene, among others.[14]
References
[edit]- ^abcdefghijklHartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA (September 2020)."The molecular virology of coronaviruses".The Journal of Biological Chemistry.295(37): 12910–12934.doi:10.1074/jbc.REV120.013930.PMC7489918.PMID32661197.
- ^abcdefghV'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V (March 2021)."Coronavirus biology and replication: implications for SARS-CoV-2".Nature Reviews. Microbiology.19(3): 155–170.doi:10.1038/s41579-020-00468-6.PMC7592455.PMID33116300.
- ^abcdefghPosthuma CC, Te Velthuis AJ, Snijder EJ (April 2017)."Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes".Virus Research.234:58–73.doi:10.1016/j.virusres.2017.01.023.PMC7114556.PMID28174054.
- ^abcdefghGulyaeva AA, Gorbalenya AE (January 2021)."A nidovirus perspective on SARS-CoV-2".Biochemical and Biophysical Research Communications.538:24–34.doi:10.1016/j.bbrc.2020.11.015.PMC7664520.PMID33413979.
- ^abWang D, Jiang A, Feng J, Li G, Guo D, Sajid M, et al. (May 2021)."The SARS-CoV-2 subgenome landscape and its novel regulatory features".Molecular Cell.81(10): 2135–2147.e5.doi:10.1016/j.molcel.2021.02.036.PMC7927579.PMID33713597.
- ^Irigoyen N, Firth AE, Jones JD, Chung BY, Siddell SG, Brierley I (February 2016)."High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling".PLOS Pathogens.12(2): e1005473.doi:10.1371/journal.ppat.1005473.PMC4769073.PMID26919232.
- ^Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, et al. (January 2021)."The coding capacity of SARS-CoV-2".Nature.589(7840): 125–130.Bibcode:2021Natur.589..125F.doi:10.1038/s41586-020-2739-1.PMID32906143.S2CID221624633.
- ^Smith EC, Denison MR (5 December 2013)."Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity".PLOS Pathogens.9(12): e1003760.doi:10.1371/journal.ppat.1003760.PMC3857799.PMID24348241.
- ^abcOgando NS, Ferron F, Decroly E, Canard B, Posthuma CC, Snijder EJ (7 August 2019)."The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity".Frontiers in Microbiology.10:1813.doi:10.3389/fmicb.2019.01813.PMC6693484.PMID31440227.
- ^Rohaim MA, El Naggar RF, Clayton E, Munir M (January 2021)."Structural and functional insights into non-structural proteins of coronaviruses".Microbial Pathogenesis.150:104641.doi:10.1016/j.micpath.2020.104641.PMC7682334.PMID33242646.
- ^abChen Y, Liu Q, Guo D (April 2020)."Emerging coronaviruses: Genome structure, replication, and pathogenesis".Journal of Medical Virology.92(4): 418–423.doi:10.1002/jmv.25681.PMC7167049.PMID31967327.
- ^Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE (November 2018)."A planarian nidovirus expands the limits of RNA genome size".PLOS Pathogens.14(11): e1007314.doi:10.1371/journal.ppat.1007314.PMC6211748.PMID30383829.S2CID53872740.
- ^Koyama T, Platt D, Parida L (July 2020)."Variant analysis of SARS-CoV-2 genomes".Bulletin of the World Health Organization.98(7): 495–504.doi:10.2471/BLT.20.253591.PMC7375210.PMID32742035.
- ^Richardson, Robin (August 22, 2021)."Open Wide".The Marshall News Messenger.pp. A1, A2.Retrieved21 November2022.

