Major prion protein
Themajor prion protein(PrP) is encoded in the human body by thePRNPgenealso known asCD230(cluster of differentiation230).[5][6][7][8]Expression of theproteinis most predominant in thenervous systembut occurs in many other tissues throughout the body.[9][10][11]
The protein can exist in multipleisoforms:the normalPrPCform, and theprotease-resistant form designatedPrPRessuch as the disease-causingPrPSc(scrapie) and an isoform located inmitochondria.Themisfoldedversion PrPScis associated with a variety ofcognitive disordersandneurodegenerativediseases such as in animals:ovinescrapie,bovine spongiform encephalopathy(BSE, mad cow disease),feline spongiform encephalopathy,transmissible mink encephalopathy(TME),exotic ungulate encephalopathy,chronic wasting disease(CWD) which affectsdeer;and in humans:Creutzfeldt–Jakob disease(CJD),fatal familial insomnia(FFI),Gerstmann–Sträussler–Scheinker syndrome(GSS),kuru,andvariant Creutzfeldt–Jakob disease(vCJD). Similarities exist between kuru, thought to be due to human ingestion of diseased individuals, and vCJD, thought to be due to human ingestion of BSE-tainted cattle products.
Gene
[edit]
The humanPRNPgene is located on the short (p) arm ofchromosome 20between the end (terminus) of the arm and position 13, frombase pair4,615,068 to base pair 4,630,233.
Structure
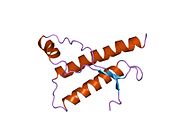
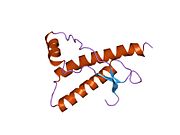
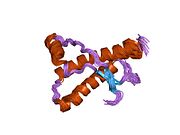
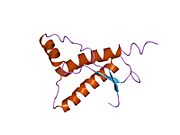
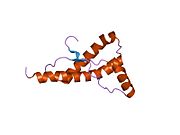
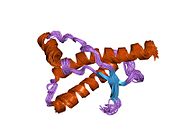
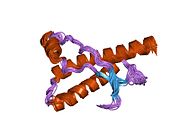
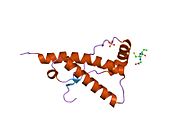
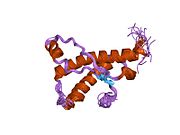
[edit]PrP is highly conserved through mammals, lending credence to application of conclusions from test animals such as mice.[12]Comparison between primates is especially similar, ranging from 92.9 to 99.6% similarity inamino acid sequences.The human protein structure consists of a globular domain with threeα-helicesand a two-strandantiparallelβ-sheet,anNH2-terminaltail, and a shortCOOH-terminaltail.[13]Aglycophosphatidylinositol(GPI) membrane anchor at the COOH-terminal tethers PrP tocell membranes,and this proves to be integral to the transmission of conformational change; secreted PrP lacking the anchor component is unaffected by the infectious isoform.[14]
The primary sequence of PrP is 253amino acidslong beforepost-translational modification.Signal sequencesin theamino- andcarboxy- terminal ends are removed posttranslationally, resulting in a mature length of 208 amino acids. For human andgolden hamsterPrP, twoglycosylatedsites exist on helices 2 and 3 atAsn181 and Asn197.MurinePrP has glycosylation sites as Asn180 and Asn196. Adisulfidebond exists betweenCys179 of the second helix and Cys214 of the third helix (human PrPCnumbering).
PrPmessenger RNAcontains apseudoknotstructure (prion pseudoknot), which is thought to be involved in regulation of PrPprotein translation.[15]
Ligand-binding
[edit]The mechanism for conformational conversion to the scrapie isoform is speculated to be an elusiveligand-protein, but, so far, no such compound has been identified. However, a large body of research has developed on candidates and their interaction with the PrPC.[16]
Copper,zinc,manganese,andnickelare confirmed PrP ligands that bind to its octarepeat region.[17]Ligand binding causes a conformational change with unknown effect. Heavy metal binding at PrP has been linked to resistance tooxidative stressarising fromheavy metal toxicity.[17][18]
PrPC(normal cellular) isoform
[edit]The precise function of PrP is not yet known. It may play a role in the transport ofioniccopper into cells from the surrounding environment. Researchers have also proposed roles for PrP in cell signaling or in the formation ofsynapses.[19]PrPCattaches to the outer surface of thecell membraneby aglycosylphosphatidylinositolanchor at itsC-terminalSer231.
Prion proteincontains fiveoctapeptiderepeats with sequence PHGGGWGQ (though the first repeat has the slightly modified,histidine-deficient sequence PQGGGGWGQ). This is thought to generate a copper-binding domainvia nitrogen atoms in the histidineimidazoleside-chainsand deprotonatedamidenitrogens from the 2nd and 3rd glycines in the repeat. The ability to bind copper is, therefore,pH-dependent.NMRshows copper binding results in aconformationalchange at theN-terminus.
PrPSc(scrapie) isoform
[edit]PrPScis a conformational isoform of PrPC,but this orientation tends to accumulate in compact,protease-resistant aggregates within neural tissue.[20]The abnormal PrPScisoform has a differentsecondaryandtertiary structurefrom PrPC,but identical primary sequence. Whereas PrPChas largely alpha helical and disordered domains,[21]PrPSchas no alpha helix and an amyloid fibril core composed of a stack of PrP molecules glued together by parallel in-register intermolecular beta sheets.[22][23][24]This refolding renders the PrPScisoform extremely resistant toproteolysis.
The propagation of PrPScis a topic of great interest, as its accumulation is a pathological cause ofneurodegeneration.Based on the progressive nature of spongiform encephalopathies, the predominant hypothesis posits that the change from normal PrPCis caused by the presence and interaction with PrPSc.[25]Strong support for this is taken from studies in whichPRNP-knockout mice are resistant to the introduction of PrPSc.[26]Despite widespread acceptance of the conformation conversion hypothesis, some studies mitigate claims for a direct link between PrPScandcytotoxicity.[27]
Polymorphismsat sites 136, 154, and 171 are associated with varying susceptibility to ovinescrapie.(These ovine sites correspond to human sites 133, 151, and 168.) Polymorphisms of the PrP-VRQ form and PrP-ARQ form are associated with increased susceptibility, whereas PrP-ARR is associated with resistance. The National Scrapie Plan of the UK aims to breed out these scrapie polymorphisms by increasing the frequency of the resistant allele.[28]However, PrP-ARR polymorphisms are susceptible to atypical scrapie, so this may prove unfruitful.
Function
[edit]Nervous system
[edit]The strong association to neurodegenerative diseases raises many questions of the function of PrP in the brain. A common approach is using PrP-knockout andtransgenicmice to investigate deficiencies and differences.[29]Initial attempts produced two strains of PrP-null mice that show no physiological or developmental differences when subjected to an array of tests. However, more recent strains have shown significant cognitive abnormalities.[16]
As the null mice age, a marked loss ofPurkinje cellsin thecerebellumresults in decreased motor coordination. However, this effect is not a direct result of PrP's absence, and rather arises from increasedDoppelgene expression.[30]Other observed differences include reduced stress response and increased exploration of novel environments.[31][32]
Circadian rhythmis altered in null mice.[11]Fatal familial insomniais thought to be the result of a point mutation inPRNPat codon 178, which corroborates PrP's involvement in sleep-wake cycles.[33]In addition, circadian regulation has been demonstrated in PrP mRNA, which cycles regularly with day-night.[34]
Memory
[edit]While null mice exhibit normal learning ability andshort-term memory,long-term memoryconsolidation deficits have been demonstrated. As withataxia,this is attributable to Doppel gene expression. However,spatial learning,a predominantly hippocampal-function, is decreased in the null mice and can be recovered with the reinstatement of PrP in neurons; this indicates that loss of PrP function is the cause.[35][36]The interaction of hippocampal PrP withlaminin(LN) is pivotal in memory processing and is likely modulated by thekinasesPKA and ERK1/2.[37][38]
Further support for PrP's role in memory formation is derived from several population studies. A test of healthy young humans showed increased long-term memory ability associated with an MM or MV genotype when compared to VV.[39]Down syndromepatients with a singlevalinesubstitution have been linked to earlier cognitive decline.[40]SeveralpolymorphismsinPRNPhave been linked with cognitive impairment in the elderly as well as earlier cognitive decline.[41][42][43]All of these studies investigated differences in codon 129, indicating its importance in the overall functionality of PrP, in particular with regard to memory.
Neurons and synapses
[edit]PrP is present in both the pre- and post-synaptic compartments, with the greatest concentration in the pre-synaptic portion.[44]Considering this and PrP's suite of behavioral influences, the neural cell functions and interactions are of particular interest. Based on the copper ligand, one proposed function casts PrP as a copper buffer for thesynaptic cleft.In this role, the protein could serve as either a copperhomeostasismechanism, a calcium modulator, or a sensor for copper or oxidative stress.[45]Loss of PrP function has been linked tolong-term potentiation(LTP). This effect can be positive or negative and is due to changes in neuronal excitability and synaptic transmission in thehippocampus.[46][47]
Some research indicates PrP involvement in neuronal development, differentiation, andneuriteoutgrowth. The PrP-activated signal transduction pathway is associated with axon and dendritic outgrowth with a series of kinases.[27][48]
Immune system
[edit]Though most attention is focused on PrP's presence in the nervous system, it is also abundant in immune system tissue. PrP immune cells include hematopoietic stem cells, mature lymphoid and myeloid compartments, and certainlymphocytes;also, it has been detected innatural killer cells,platelets,andmonocytes.T cellactivation is accompanied by a strong up-regulation of PrP, though it is not requisite. The lack of immunoresponse totransmissible spongiform encephalopathies(TSE), neurodegenerative diseases caused by prions, could stem from the tolerance for PrPSc.[49]
Muscles, liver, and pituitary
[edit]PrP-null mice provide clues to a role in muscular physiology when subjected to aforced swimming test,which showed reduced locomotor activity. Aging mice with an overexpression of PRNP showed significant degradation of muscle tissue.
Though present, very low levels of PrP exist in the liver and could be associated with liver fibrosis. Presence in the pituitary has been shown to affect neuroendocrine function in amphibians, but little is known concerning mammalian pituitary PrP.[16]
Cellular
[edit]Varying expression of PrP through thecell cyclehas led to speculation on involvement in development. A wide range of studies has been conducted investigating the role in cell proliferation, differentiation, death, and survival.[16]Engagement of PrP has been linked to activation ofsignal transduction.
Modulation of signal transduction pathways has been demonstrated in cross-linking with antibodies and ligand-binding (hop/STI1 or copper).[16]Given the diversity of interactions, effects, and distribution, PrP has been proposed as dynamic surface protein functioning in signaling pathways. Specific sites along the protein bind other proteins, biomolecules, and metals. These interfaces allow specific sets of cells to communicate based on level of expression and the surrounding microenvironment. The anchoring on aGPI raftin thelipid bilayersupports claims of anextracellular scaffoldingfunction.[16]
Diseases caused by PrP misfolding
[edit]More than 20 mutations in thePRNPgene have been identified in people with inheritedprion diseases,which include the following:[50][51]
- Creutzfeldt–Jakob disease–glutamic acid-200 is replaced bylysinewhilevalineis present at amino acid 129
- Gerstmann–Sträussler–Scheinker syndrome– usually a change incodon102 fromprolinetoleucine[52]
- fatal familial insomnia–aspartic acid-178 is replaced byasparaginewhilemethionineis present at amino acid 129[53]
The conversion of PrPCto PrPScconformation is the mechanism of transmission of fatal, neurodegenerative transmissible spongiform encephalopathies (TSE). This can arise from genetic factors, infection from external source, or spontaneously for reasons unknown. Accumulation of PrPSccorresponds with progression of neurodegeneration and is the proposed cause. SomePRNPmutations lead to a change in singleamino acids(the building-blocks of proteins) in the prion protein. Others insert additional amino acids into the protein or cause an abnormally short protein to be made. These mutations cause the cell to make prion proteins with an abnormal structure. The abnormal protein PrPScaccumulates in the brain and destroys nerve cells, which leads to the mental and behavioral features of prion diseases.
Several other changes in thePRNPgene (called polymorphisms) do not cause prion diseases but may affect a person's risk of developing these diseases or alter the course of the disorders. Anallelethat codes for a PRNP variant, G127V, provides resistance tokuru.[54]
In addition, some prion diseases can be transmitted from external sources of PrPSc.[55]
- Scrapie– fatal neurodegenerative disease in sheep, not transmissible to humans
- Bovine spongiform encephalopathy(mad-cow disease) – fatal neurodegenerative disease in cows, which can be transmitted to humans by ingestion of brain, spinal, or digestive tract tissue of an infected cow
- Kuru– TSE in humans, transmitted via funerary cannibalism. Generally, affected family members were given, by tradition, parts of the central nervous system according to ritual when consuming deceased family members.
Alzheimer's disease
[edit]PrPCprotein is one of several cellular receptors of solubleamyloid beta(Aβ) oligomers, which are canonically implicated in causingAlzheimer's disease.[56]Theseoligomersare composed of smaller Aβ plaques, and are the most damaging to the integrity of aneuron.[56]The precise mechanism of soluble Aβ oligomers directly inducingneurotoxicityis unknown, and experimental deletion ofPRNPin animals has yielded several conflicting findings. When Aβ oligomers were injected into thecerebral ventriclesof a mouse model of Alzheimer's,PRNPdeletion did not offer protection, only anti-PrPCantibodies prevented long-term memory andspatial learningdeficits.[57][58]This would suggest either an unequal relation between PRNP and Aβ oligomer-mediatedneurodegenerationor a site-specific relational significance. In the case of direct injection of Aβ oligomers into thehippocampus,PRNP-knockout mice were found to be indistinguishable from control with respect to both neuronal death rates and measurements ofsynaptic plasticity.[56][58]It was further found that Aβ-oligomers bind to PrPCat thepostsynaptic density,indirectly overactivating theNMDA receptorvia theFynenzyme, resulting inexcitotoxicity.[57]Soluble Aβ oligomers also bind to PrPCat thedendritic spines,forming a complex with Fyn and excessively activatingtau,another protein implicated in Alzheimer's.[57]As the geneFYNcodes for the enzyme Fyn, FYN-knockout mice display neitherexcitotoxicevents nordendritic spine shrinkagewhen injected with Aβ oligomers.[57]In mammals, the full functional significance of PRNP remains unclear, asPRNPdeletion has been prophylactically implemented by the cattle industry without apparent harm.[56]In mice, this same deletionphenotypicallyvaries between Alzheimer's mouse lines, as hAPPJ20 mice and TgCRND8 mice show a slight increase inepilepticactivity, contributing to conflicting results when examining Alzheimer's survival rates.[56]Of note, the deletion ofPRNPin both APPswe and SEN1dE9, two othertransgenicmodels of Alzheimer's, attenuated the epilepsy-induced death phenotype seen in a subset of these animals.[56]Taken collectively, recent evidence suggests PRNP may be important for conducing the neurotoxic effects of soluble Aβ-oligomers and the emergent disease state of Alzheimer's.[56][57][58]
In humans, themethionine/valinepolymorphismatcodon129 ofPRNP(rs1799990) is most closely associated with Alzheimer's disease.[59]Variant Vallelecarriers (VV and MV) show a 13% decreased risk with respect to developing Alzheimer's compared to the methioninehomozygote(MM). However, the protective effects of variant V carriers have been found exclusively inCaucasians.The decreased risk in V allele carriers is further limited to late-onset Alzheimer's disease only (≥ 65 years).[59]PRNP can also functionally interact with polymorphisms in two other genes implicated in Alzheimer's,PSEN1andAPOE,to compound risk for both Alzheimer's andsporadic Creutzfeldt–Jakob disease.[56]Apoint mutationon codon 102 ofPRNPat least in part contributed to three separate patients' atypicalfrontotemporal dementiawithin the same family, suggesting a new phenotype forGerstmann–Sträussler–Scheinker syndrome.[56][60]The same study proposed sequencingPRNPin cases of ambiguously diagnosed dementia, as the various forms ofdementiacan prove challenging todifferentially diagnose.[60]
Research
[edit]In 2006 the production of cattle lacking PrPCform of the major prion protein (PrP) protein was reported which were resistant to prion propagation with no apparent developmental abnormalities. Besides the study of bovine products free of prion proteins another use could be so that human pharmaceuticals can be made in their blood without the danger that those products might get contaminated with the infectious agent that causes mad cow.[61][62]
Interactions
[edit]A stronginteractionexists between PrP and thecochaperoneHop(Hsp70/Hsp90organizing protein; also called STI1 (Stress-induced protein 1)).[63][64]
References
[edit]- ^abcGRCh38: Ensembl release 89: ENSG00000171867–Ensembl,May 2017
- ^abcGRCm38: Ensembl release 89: ENSMUSG00000079037–Ensembl,May 2017
- ^"Human PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^"Mouse PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^Kretzschmar HA, Stowring LE, Westaway D, Stubblebine WH, Prusiner SB, Dearmond SJ (August 1986). "Molecular cloning of a human prion protein cDNA".DNA.5(4): 315–324.doi:10.1089/dna.1986.5.315.PMID3755672.
- ^Sparkes RS, Simon M, Cohn VH, Fournier RE, Lem J, Klisak I, et al. (October 1986)."Assignment of the human and mouse prion protein genes to homologous chromosomes".Proceedings of the National Academy of Sciences of the United States of America.83(19): 7358–7362.Bibcode:1986PNAS...83.7358S.doi:10.1073/pnas.83.19.7358.PMC386716.PMID3094007.
- ^Liao YC, Lebo RV, Clawson GA, Smuckler EA (July 1986). "Human prion protein cDNA: molecular cloning, chromosomal mapping, and biological implications".Science.233(4761): 364–367.Bibcode:1986Sci...233..364L.doi:10.1126/science.3014653.PMID3014653.
- ^Robakis NK, Devine-Gage EA, Jenkins EC, Kascsak RJ, Brown WT, Krawczun MS, Silverman WP (October 1986). "Localization of a human gene homologous to the PrP gene on the p arm of chromosome 20 and detection of PrP-related antigens in normal human brain".Biochemical and Biophysical Research Communications.140(2): 758–765.doi:10.1016/0006-291X(86)90796-5.PMID2877664.
- ^Prusiner SB (May 2001)."Shattuck lecture--neurodegenerative diseases and prions".The New England Journal of Medicine.344(20): 1516–1526.doi:10.1056/NEJM200105173442006.PMID11357156.
- ^Weissmann C (November 2004). "The state of the prion".Nature Reviews. Microbiology.2(11): 861–871.doi:10.1038/nrmicro1025.PMID15494743.S2CID20992257.
- ^abZomosa-Signoret V, Arnaud JD, Fontes P, Alvarez-Martinez MT, Liautard JP (2008)."Physiological role of the cellular prion protein".Veterinary Research.39(4): 9.doi:10.1051/vetres:2007048.PMID18073096.
- ^Damberger FF, Christen B, Pérez DR, Hornemann S, Wüthrich K (October 2011)."Cellular prion protein conformation and function".Proceedings of the National Academy of Sciences of the United States of America.108(42): 17308–17313.Bibcode:2011PNAS..10817308D.doi:10.1073/pnas.1106325108.PMC3198368.PMID21987789.
- ^Schätzl HM, Da Costa M, Taylor L, Cohen FE, Prusiner SB (January 1995). "Prion protein gene variation among primates".Journal of Molecular Biology.245(4): 362–374.doi:10.1006/jmbi.1994.0030.PMID7837269.
- ^Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, et al. (June 2005). "Anchorless prion protein results in infectious amyloid disease without clinical scrapie".Science.308(5727): 1435–1439.Bibcode:2005Sci...308.1435C.CiteSeerX10.1.1.401.781.doi:10.1126/science.1110837.PMID15933194.S2CID10064966.
- ^Barrette I, Poisson G, Gendron P, Major F (February 2001)."Pseudoknots in prion protein mRNAs confirmed by comparative sequence analysis and pattern searching".Nucleic Acids Research.29(3): 753–758.doi:10.1093/nar/29.3.753.PMC30388.PMID11160898.
- ^abcdefLinden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR (April 2008). "Physiology of the prion protein".Physiological Reviews.88(2): 673–728.doi:10.1152/physrev.00007.2007.PMID18391177.
- ^abPrčina M, Kontseková E, Novák M (June 2015)."Prion protein prevents heavy metals overloading of cells and thus protects them against their toxicity".Acta Virologica.59(2): 179–184.doi:10.4149/av_2015_02_179.PMID26104335.
- ^Brown DR, Clive C, Haswell SJ (January 2001)."Antioxidant activity related to copper binding of native prion protein".Journal of Neurochemistry.76(1): 69–76.doi:10.1046/j.1471-4159.2001.00009.x.PMID11145979.S2CID45647133.
- ^Kanaani J, Prusiner SB, Diacovo J, Baekkeskov S, Legname G (December 2005)."Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro".Journal of Neurochemistry.95(5): 1373–1386.doi:10.1111/j.1471-4159.2005.03469.x.PMID16313516.S2CID24329326.
- ^Ross CA, Poirier MA (July 2004). "Protein aggregation and neurodegenerative disease".Nature Medicine.10(7 Suppl): S10–S17.doi:10.1038/nm1066.PMID15272267.S2CID205383483.
- ^Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K (August 1997)."NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231)"(PDF).FEBS Letters.413(2): 282–288.Bibcode:1997FEBSL.413..282R.doi:10.1016/S0014-5793(97)00920-4.PMID9280298.S2CID39791520.
- ^Kraus A, Hoyt F, Schwartz CL, Hansen B, Artikis E, Hughson AG, et al. (November 2021). "High-resolution structure and strain comparison of infectious mammalian prions".Molecular Cell.81(21): 4540–4551.e6.doi:10.1016/j.molcel.2021.08.011.PMID34433091.
- ^Manka SW, Zhang W, Wenborn A, Betts J, Joiner S, Saibil HR, et al. (July 2022)."2.7 Å cryo-EM structure of ex vivo RML prion fibrils".Nature Communications.13(1): 4004.Bibcode:2022NatCo..13.4004M.doi:10.1038/s41467-022-30457-7.PMC9279362.PMID35831275.
- ^Hoyt F, Standke HG, Artikis E, Schwartz CL, Hansen B, Li K, et al. (July 2022)."Cryo-EM structure of anchorless RML prion reveals variations in shared motifs between distinct strains".Nature Communications.13(1): 4005.Bibcode:2022NatCo..13.4005H.doi:10.1038/s41467-022-30458-6.PMC9279418.PMID35831291.
- ^Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J (February 2011). "Prion propagation and toxicity in vivo occur in two distinct mechanistic phases".Nature.470(7335): 540–542.Bibcode:2011Natur.470..540S.doi:10.1038/nature09768.PMID21350487.S2CID4399936.
- ^Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C (July 1993)."Mice devoid of PrP are resistant to scrapie".Cell.73(7): 1339–1347.doi:10.1016/0092-8674(93)90360-3.PMID8100741.
- ^abAguzzi A, Baumann F, Bremer J (2008). "The prion's elusive reason for being".Annual Review of Neuroscience.31:439–477.doi:10.1146/annurev.neuro.31.060407.125620.PMID18558863.
- ^Atkinson M (October 2001). "National scrapie plan".The Veterinary Record.149(15): 462.PMID11688751.
- ^Weissmann C, Flechsig E (2003)."PrP knock-out and PrP transgenic mice in prion research".British Medical Bulletin.66:43–60.doi:10.1093/bmb/66.1.43.PMID14522848.
- ^Katamine S, Nishida N, Sugimoto T, Noda T, Sakaguchi S, Shigematsu K, et al. (December 1998). "Impaired motor coordination in mice lacking prion protein".Cellular and Molecular Neurobiology.18(6): 731–742.doi:10.1023/A:1020234321879.PMID9876879.S2CID23409873.
- ^Nico PB, de-Paris F, Vinadé ER, Amaral OB, Rockenbach I, Soares BL, et al. (July 2005). "Altered behavioural response to acute stress in mice lacking cellular prion protein".Behavioural Brain Research.162(2): 173–181.doi:10.1016/j.bbr.2005.02.003.PMID15970215.S2CID37511702.
- ^Roesler R, Walz R, Quevedo J, de-Paris F, Zanata SM, Graner E, et al. (August 1999). "Normal inhibitory avoidance learning and anxiety, but increased locomotor activity in mice devoid of PrP(C)".Brain Research. Molecular Brain Research.71(2): 349–353.doi:10.1016/S0169-328X(99)00193-X.PMID10521590.
- ^Medori R, Tritschler HJ, LeBlanc A, Villare F, Manetto V, Chen HY, et al. (February 1992)."Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene".The New England Journal of Medicine.326(7): 444–449.doi:10.1056/NEJM199202133260704.PMC6151859.PMID1346338.
- ^Cagampang FR, Whatley SA, Mitchell AL, Powell JF, Campbell IC, Coen CW (1999). "Circadian regulation of prion protein messenger RNA in the rat forebrain: a widespread and synchronous rhythm".Neuroscience.91(4): 1201–1204.doi:10.1016/S0306-4522(99)00092-5.PMID10391428.S2CID42892475.
- ^Criado JR, Sánchez-Alavez M, Conti B, Giacchino JL, Wills DN, Henriksen SJ, et al. (2005). "Mice devoid of prion protein have cognitive deficits that are rescued by reconstitution of PrP in neurons".Neurobiology of Disease.19(1–2): 255–265.doi:10.1016/j.nbd.2005.01.001.PMID15837581.S2CID2618712.
- ^Balducci C, Beeg M, Stravalaci M, Bastone A, Sclip A, Biasini E, et al. (February 2010)."Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein".Proceedings of the National Academy of Sciences of the United States of America.107(5): 2295–2300.Bibcode:2010PNAS..107.2295B.doi:10.1073/pnas.0911829107.PMC2836680.PMID20133875.
- ^Coitinho AS, Freitas AR, Lopes MH, Hajj GN, Roesler R, Walz R, et al. (December 2006). "The interaction between prion protein and laminin modulates memory consolidation".The European Journal of Neuroscience.24(11): 3255–3264.doi:10.1111/j.1460-9568.2006.05156.x.PMID17156386.S2CID17164351.
- ^Shorter J, Lindquist S (June 2005). "Prions as adaptive conduits of memory and inheritance".Nature Reviews. Genetics.6(6): 435–450.doi:10.1038/nrg1616.PMID15931169.S2CID5575951.
- ^Papassotiropoulos A, Wollmer MA, Aguzzi A, Hock C, Nitsch RM, de Quervain DJ (August 2005)."The prion gene is associated with human long-term memory".Human Molecular Genetics.14(15): 2241–2246.doi:10.1093/hmg/ddi228.PMID15987701.
- ^Del Bo R, Comi GP, Giorda R, Crimi M, Locatelli F, Martinelli-Boneschi F, et al. (June 2003). "The 129 codon polymorphism of the prion protein gene influences earlier cognitive performance in Down syndrome subjects".Journal of Neurology.250(6): 688–692.doi:10.1007/s00415-003-1057-5.PMID12796830.S2CID21049364.
- ^Berr C, Richard F, Dufouil C, Amant C, Alperovitch A, Amouyel P (September 1998). "Polymorphism of the prion protein is associated with cognitive impairment in the elderly: the EVA study".Neurology.51(3): 734–737.doi:10.1212/wnl.51.3.734.PMID9748018.S2CID11352163.
- ^Croes EA, Dermaut B, Houwing-Duistermaat JJ, Van den Broeck M, Cruts M, Breteler MM, et al. (August 2003). "Early cognitive decline is associated with prion protein codon 129 polymorphism".Annals of Neurology.54(2): 275–276.doi:10.1002/ana.10658.PMID12891686.S2CID31538672.
- ^Kachiwala SJ, Harris SE, Wright AF, Hayward C, Starr JM, Whalley LJ, Deary IJ (September 2005). "Genetic influences on oxidative stress and their association with normal cognitive ageing".Neuroscience Letters.386(2): 116–120.doi:10.1016/j.neulet.2005.05.067.PMID16023289.S2CID23642220.
- ^Herms J, Tings T, Gall S, Madlung A, Giese A, Siebert H, et al. (October 1999)."Evidence of presynaptic location and function of the prion protein".The Journal of Neuroscience.19(20): 8866–8875.doi:10.1523/JNEUROSCI.19-20-08866.1999.PMC6782778.PMID10516306.
- ^Kardos J, Kovács I, Hajós F, Kálmán M, Simonyi M (August 1989). "Nerve endings from rat brain tissue release copper upon depolarization. A possible role in regulating neuronal excitability".Neuroscience Letters.103(2): 139–144.doi:10.1016/0304-3940(89)90565-X.PMID2549468.S2CID24917999.
- ^Bailey CH, Kandel ER, Si K (September 2004)."The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth".Neuron.44(1): 49–57.doi:10.1016/j.neuron.2004.09.017.PMID15450159.S2CID2637074.
- ^Barco A, Bailey CH, Kandel ER (June 2006)."Common molecular mechanisms in explicit and implicit memory".Journal of Neurochemistry.97(6): 1520–1533.doi:10.1111/j.1471-4159.2006.03870.x.PMID16805766.S2CID26307975.
- ^Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM (February 2009)."Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers".Nature.457(7233): 1128–1132.Bibcode:2009Natur.457.1128L.doi:10.1038/nature07761.PMC2748841.PMID19242475.
- ^Isaacs JD, Jackson GS, Altmann DM (October 2006)."The role of the cellular prion protein in the immune system".Clinical and Experimental Immunology.146(1): 1–8.doi:10.1111/j.1365-2249.2006.03194.x.PMC1809729.PMID16968391.
- ^Castilla J, Hetz C, Soto C (June 2004). "Molecular mechanisms of neurotoxicity of pathological prion protein".Current Molecular Medicine.4(4): 397–403.doi:10.2174/1566524043360654.PMID15354870.
- ^Kovács GG, Trabattoni G, Hainfellner JA, Ironside JW, Knight RS, Budka H (November 2002). "Mutations of the prion protein gene phenotypic spectrum".Journal of Neurology.249(11): 1567–1582.doi:10.1007/s00415-002-0896-9.PMID12420099.S2CID22688729.
- ^Collins S, McLean CA, Masters CL (September 2001). "Gerstmann-Sträussler-Scheinker syndrome,fatal familial insomnia, and kuru: a review of these less common human transmissible spongiform encephalopathies".Journal of Clinical Neuroscience.8(5): 387–397.doi:10.1054/jocn.2001.0919.PMID11535002.S2CID31976428.
- ^Montagna P, Gambetti P, Cortelli P, Lugaresi E (March 2003). "Familial and sporadic fatal insomnia".The Lancet. Neurology.2(3): 167–176.doi:10.1016/S1474-4422(03)00323-5.PMID12849238.S2CID20822956.
- ^Mead S, Whitfield J, Poulter M, Shah P, Uphill J, Campbell T, et al. (November 2009)."A novel protective prion protein variant that colocalizes with kuru exposure".The New England Journal of Medicine.361(21): 2056–2065.doi:10.1056/NEJMoa0809716.PMID19923577.
- "Brain disease 'resistance gene' evolves in Papua New Guinea community; could offer insights into CJD".ScienceDaily(Press release). November 21, 2009.
- ^Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho JH, Petritis B, et al. (2009)."A systems approach to prion disease".Molecular Systems Biology.5(1): 252.doi:10.1038/msb.2009.10.PMC2671916.PMID19308092.
- ^abcdefghiLaurén J (2014). "Cellular prion protein as a therapeutic target in Alzheimer's disease".Journal of Alzheimer's Disease.38(2): 227–244.doi:10.3233/JAD-130950.PMID23948943.
- ^abcdeZhou J, Liu B (May 2013)."Alzheimer's disease and prion protein".Intractable & Rare Diseases Research.2(2): 35–44.doi:10.5582/irdr.2013.v2.2.35.PMC4204584.PMID25343100.
- ^abcLaurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM (February 2009)."Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers".Nature.457(7233): 1128–1132.Bibcode:2009Natur.457.1128L.doi:10.1038/nature07761.PMC2748841.PMID19242475.
- ^abHe J, Li X, Yang J, Huang J, Fu X, Zhang Y, Fan H (March 2013). "The association between the methionine/valine (M/V) polymorphism (rs1799990) in the PRNP gene and the risk of Alzheimer disease: an update by meta-analysis".Journal of the Neurological Sciences.326(1–2): 89–95.doi:10.1016/j.jns.2013.01.020.PMID23399523.S2CID31070331.
- ^abGiovagnoli AR, Di Fede G, Aresi A, Reati F, Rossi G, Tagliavini F (December 2008). "Atypical frontotemporal dementia as a new clinical phenotype of Gerstmann-Straussler-Scheinker disease with the PrP-P102L mutation. Description of a previously unreported Italian family".Neurological Sciences.29(6): 405–410.doi:10.1007/s10072-008-1025-z.PMID19030774.S2CID20553167.
- ^Weiss R (1 January 2007)."Scientists Announce Mad Cow Breakthrough".The Washington Post.Retrieved1 January2007.
- ^Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, et al. (January 2007)."Production of cattle lacking prion protein".Nature Biotechnology.25(1): 132–138.doi:10.1038/nbt1271.PMC2813193.PMID17195841.
- ^Americo TA, Chiarini LB, Linden R (June 2007). "Signaling induced by hop/STI-1 depends on endocytosis".Biochemical and Biophysical Research Communications.358(2): 620–625.doi:10.1016/j.bbrc.2007.04.202.PMID17498662.
- ^Zanata SM, Lopes MH, Mercadante AF, Hajj GN, Chiarini LB, Nomizo R, et al. (July 2002)."Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection".The EMBO Journal.21(13): 3307–3316.doi:10.1093/emboj/cdf325.PMC125391.PMID12093732.
External links
[edit]- PRNP (PrP) gene atGeneCard
- PRNP+protein,+humanat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
- Susan Lindquist's Seminar: "The Surprising World of Prion Biology"