Paracrine signaling
Incellular biology,paracrine signalingis a form ofcell signaling,a type ofcellular communicationin which acellproduces a signal to induce changes in nearby cells, altering the behaviour of those cells. Signaling molecules known asparacrine factorsdiffuseover a relatively short distance (local action), as opposed to cell signaling byendocrine factors,hormoneswhich travel considerably longer distances via thecirculatory system;juxtacrine interactions;andautocrine signaling.Cells that produce paracrine factors secrete them into the immediateextracellularenvironment. Factors then travel to nearby cells in which the gradient of factor received determines the outcome. However, the exact distance that paracrine factors can travel is not certain.
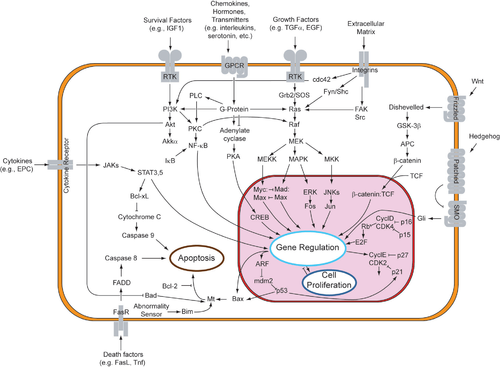
Although paracrine signaling elicits a diverse array of responses in the induced cells, most paracrine factors utilize a relatively streamlined set ofreceptorsand pathways. In fact, differentorgansin the body - even between different species - are known to utilize a similar sets of paracrine factors in differential development.[1]The highly conserved receptors and pathways can be organized into four major families based on similar structures:fibroblast growth factor(FGF) family,Hedgehogfamily,Wntfamily, andTGF-β superfamily.Binding of a paracrine factor to its respective receptor initiatessignal transductioncascades, eliciting different responses.
Paracrine factors induce competent responders[edit]
In order for paracrine factors to successfully induce a response in the receiving cell, that cell must have the appropriate receptors available on the cell membrane to receive the signals, also known as beingcompetent.Additionally, the responding cell must also have the ability to be mechanistically induced.
Fibroblast growth factor (FGF) family[edit]
Although the FGF family of paracrine factors has a broad range of functions, major findings support the idea that they primarily stimulate proliferation and differentiation.[2][3]To fulfill many diverse functions, FGFs can be alternatively spliced or even have different initiation codons to create hundreds of different FGFisoforms.[4]
One of the most important functions of the FGF receptors (FGFR) is in limb development. This signaling involves nine differentalternatively splicedisoformsof the receptor.[5]Fgf8 andFgf10 are two of the critical players in limb development. In the forelimb initiation and limb growth in mice, axial (lengthwise) cues from the intermediatemesodermproducesTbx5, which subsequently signals to the samemesodermto produceFgf10.Fgf10 then signals to theectodermto begin production ofFgf8, which also stimulates the production ofFgf10. Deletion ofFgf10 results in limbless mice.[6]
Additionally, paracrine signaling of Fgf is essential in the developing eye of chicks. Thefgf8mRNAbecomes localized in what differentiates into the neuralretinaof theoptic cup.These cells are in contact with the outer ectoderm cells, which will eventually become the lens.[4]
Phenotypeand survival of mice after knockout of some FGFR genes:[5]
| FGFR Knockout Gene | Survival | Phenotype |
|---|---|---|
| Fgf1 | Viable | Unclear |
| Fgf3 | Viable | Inner ear, skeletal (tail) differentiation |
| Fgf4 | Lethal | Inner cell mass proliferation |
| Fgf8 | Lethal | Gastrulationdefect, CNS development, limb development |
| Fgf10 | Lethal | Development of multiple organs (including limbs, thymus, pituitary) |
| Fgf17 | Viable | Cerebellar Development |
Receptor tyrosine kinase (RTK) pathway[edit]
Paracrine signaling throughfibroblast growth factorsand its respective receptors utilizes the receptortyrosinepathway. This signaling pathway has been highly studied, usingDrosophilaeyes and human cancers.[7]
Binding of FGF to FGFRphosphorylatesthe idlekinaseand activates the RTK pathway. This pathway begins at the cell membrane surface, where aligandbinds to its specific receptor. Ligands that bind to RTKs includefibroblast growth factors,epidermal growth factors, platelet-derived growth factors, andstem cell factor.[7]This dimerizes the transmembrane receptor to another RTK receptor, which causes the autophosphorylation and subsequentconformational changeof thehomodimerizedreceptor. This conformational change activates the dormant kinase of each RTK on the tyrosine residue. Due to the fact that the receptor spans across the membrane from the extracellular environment, through thelipid bilayer,and into thecytoplasm,the binding of the receptor to the ligand also causes the trans phosphorylation of the cytoplasmic domain of the receptor.[8]
Anadaptor protein(such as SOS) recognizes the phosphorylated tyrosine on the receptor. This protein functions as a bridge which connects the RTK to an intermediate protein (such as GNRP), starting the intracellular signaling cascade. In turn, the intermediate protein stimulates GDP-bound Ras to the activated GTP-bound Ras. GAP eventually returns Ras to its inactive state. Activation ofRashas the potential to initiate three signaling pathways downstream of Ras: Ras→Raf→MAP kinase pathway, PI3 kinase pathway, and Ral pathway. Each pathway leads to the activation of transcription factors which enter the nucleus to alter gene expression.[9]

RTK receptor and cancer[edit]
Paracrine signaling of growth factors between nearby cells has been shown to exacerbatecarcinogenesis.In fact, mutant forms of a single RTK may play a causal role in very different types of cancer. The Kitproto-oncogeneencodes a tyrosine kinase receptor whose ligand is a paracrine protein called stem cell factor (SCF), which is important inhematopoiesis(formation of cells in blood).[10]The Kit receptor and related tyrosine kinase receptors actually are inhibitory and effectively suppresses receptor firing. Mutant forms of the Kit receptor, which fire constitutively in a ligand-independent fashion, are found in a diverse array of cancerous malignancies.[11]
RTK pathway and cancer[edit]
Research onthyroid cancerhas elucidated the theory that paracrine signaling may aid in creating tumor microenvironments.Chemokinetranscription is upregulated when Ras is in the GTP-bound state. The chemokines are then released from the cell, free to bind to another nearby cell. Paracrine signaling between neighboring cells creates this positive feedback loop. Thus, the constitutive transcription of upregulated proteins form ideal environments for tumors to arise.[12]Effectively, multiple bindings of ligands to the RTK receptors overstimulates the Ras-Raf-MAPK pathway, whichoverexpressesthemitogenicand invasive capacity of cells.[13]
JAK-STAT pathway[edit]
In addition to RTK pathway,fibroblast growth factorscan also activate theJAK-STAT signaling pathway.Instead of carrying covalently associated tyrosine kinase domains, Jak-STAT receptors form noncovalent complexes with tyrosine kinases of the Jak (Janus kinase) class. These receptors bind are forerythropoietin(important forerythropoiesis),thrombopoietin(important forplateletformation), andinterferon(important for mediating immune cell function).[14]
After dimerization of the cytokine receptors following ligand binding, the JAKs transphosphorylate each other. The resulting phosphotyrosines attract STAT proteins. The STAT proteins dimerize and enter the nucleus to act astranscription factorsto alter gene expression.[14]In particular, the STATs transcribe genes that aid in cell proliferation and survival – such as myc.[15]
Phenotype and survival of mice after knockout of some JAK or STAT genes:[16]
| Knockout Gene | Survival | Phenotype |
|---|---|---|
| Jak1 | Lethal | Neurologic Deficits |
| Jak2 | Lethal | Failure in erythropoiesis |
| Stat1 | Viable | Human dwarfism andcraniosynostosissyndromes |
| Stat3 | Lethal | Tissue specific phenotypes |
| Stat4 | Viable | defective IL-12-driven Th1 differentiation, increased susceptibility to intracellular pathogens |
Aberrant JAK-STAT pathway and bone mutations[edit]
The JAK-STAT signaling pathway is instrumental in the development of limbs, specifically in its ability to regulate bone growth through paracrine signaling of cytokines. However, mutations in this pathway have been implicated in severe forms of dwarfism:thanatophoric dysplasia(lethal) andachondroplasicdwarfism (viable).[17]This is due to a mutation in aFgfgene, causing a premature and constitutive activation of theStat1transcription factor.Chondrocytecell division is prematurely terminated, resulting in lethal dwarfism. Rib and limb bone growth plate cells are not transcribed. Thus, the inability of the rib cage to expand prevents the newborn's breathing.[18]
JAK-STAT pathway and cancer[edit]
Research on paracrine signaling through the JAK-STAT pathway revealed its potential in activating invasive behavior of ovarianepithelial cells.This epithelial tomesenchymaltransition is highly evident inmetastasis.[19]Paracrine signaling through the JAK-STAT pathway is necessary in the transition from stationary epithelial cells to mobile mesenchymal cells, which are capable of invading surrounding tissue. Only the JAK-STAT pathway has been found to induce migratory cells.[20]
Hedgehog family[edit]
TheHedgehog protein familyis involved in induction of cell types and the creation of tissue boundaries and patterning and are found in all bilateral organisms. Hedgehog proteins were first discovered and studied inDrosophila.Hedgehog proteins produce key signals for the establishment of limb andbody planof fruit flies as well ashomeostasisof adult tissues, involved in lateembryogenesisandmetamorphosis.At least three "Drosophila" hedgehoghomologshave been found in vertebrates: sonic hedgehog, desert hedgehog, and Indian hedgehog. Sonic hedgehog (SHH) has various roles in vertebrae development, mediating signaling and regulating the organization of central nervous system, limb, andsomitepolarity.Desert hedgehog (DHH) is expressed in theSertoli cellsinvolved inspermatogenesis.Indian hedgehog (IHH) is expressed in the gut and cartilage, important in postnatal bone growth.[21][22][23]
Hedgehog signaling pathway[edit]


Members of the Hedgehog protein family act by binding to atransmembrane"Patched"receptor, which is bound to the"Smoothened"protein, by which the Hedgehog signal can betransduced.In the absence of Hedgehog, the Patched receptor inhibits Smoothened action. Inhibition of Smoothened causes theCubitus interruptus(Ci), Fused, and Cos protein complex attached to microtubules to remain intact. In this conformation, the Ci protein is cleaved so that a portion of the protein is allowed to enter the nucleus and act as a transcriptionalrepressor.In the presence of Hedgehog, Patched no longer inhibits Smoothened. Then active Smoothened protein is able to inhibitPKAand Slimb, so that the Ci protein is not cleaved. This intact Ci protein can enter the nucleus, associate with CPB protein and act as a transcriptionalactivator,inducing the expression of Hedgehog-response genes.[23][24][25]
Hedgehog signaling pathway and cancer[edit]
The Hedgehog Signaling pathway is critical in proper tissue patterning and orientation during normal development of most animals. Hedgehog proteins inducecell proliferationin certain cells and differentiations in others. Aberrant activation of the Hedgehog pathway has been implicated in several types ofcancers,Basal Cell Carcinomain particular. This uncontrolled activation of the Hedgehog proteins can be caused by mutations to the signal pathway, which would beligandindependent, or a mutation that causesoverexpressionof the Hedgehog protein, which would be ligand dependent. In addition, therapy-induced Hedgehog pathway activation has been shown to be necessary for progression of Prostate Cancer tumors afterandrogen deprivation therapy.[26]This connection between the Hedgehog signaling pathway and human cancers may provide for the possible of therapeutic intervention as treatment for such cancers. The Hedgehog signaling pathway is also involved in normal regulation ofstem-cellpopulations, and required for normal growth and regeneration of damaged organs. This may provide another possible route fortumorigenesisvia the Hedgehog pathway.[27][28][29]
Wnt family[edit]

TheWnt proteinfamily includes a large number ofcysteine-richglycoproteins.The Wnt proteins activatesignal transductioncascades via three different pathways, the canonicalWnt pathway,the noncanonicalplanar cell polarity (PCP) pathway,and the noncanonical Wnt/Ca2+pathway. Wnt proteins appear to control a wide range of developmental processes and have been seen as necessary for control ofspindleorientation, cell polarity, cadherin mediated adhesion, and early development of embryos in many different organisms. Current research has indicated that deregulation of Wnt signaling plays a role in tumor formation, because at a cellular level, Wnt proteins often regulatedcell proliferation,cell morphology, cellmotility,and cell fate.[30]
The canonical Wnt signaling pathway[edit]

In thecanonical pathway,Wnt proteins binds to its transmembrane receptor of theFrizzledfamily of proteins. The binding of Wnt to a Frizzled protein activates theDishevelledprotein. In its active state the Dishevelled protein inhibits the activity of the glycogen synthase kinase 3 (GSK3) enzyme. Normally active GSK3 prevents the dissociation of β-catenin to theAPCprotein, which results inβ-catenindegradation. Thus inhibited GSK3, allows β-catenin to dissociate from APC, accumulate, and travel to nucleus. In the nucleus β-catenin associates with Lef/Tcftranscription factor,which is already working on DNA as a repressor, inhibiting the transcription of the genes it binds. Binding of β-catenin to Lef/Tcf works as a transcription activator, activating the transcription of the Wnt-responsive genes.[31][32][33]
The noncanonical Wnt signaling pathways[edit]
The noncanonical Wnt pathways provide a signal transduction pathway for Wnt that does not involveβ-catenin.In the noncanonical pathways, Wnt affects theactinandmicrotubularcytoskeletonas well asgene transcription.
The noncanonical planar cell polarity (PCP) pathway[edit]

The noncanonical PCP pathway regulates cellmorphology,division,andmovement.Once again Wnt proteins binds to and activates Frizzled so that Frizzled activates a Dishevelled protein that is tethered to the plasma membrane through aPrickle proteinand transmembrane Stbm protein. The active Dishevelled activates RhoAGTPasethrough Dishevelled associated activator ofmorphogenesis 1(Daam1) and theRac protein.Active RhoA is able to induce cytoskeleton changes by activating Roh-associated kinase (ROCK) and affect gene transcription directly. Active Rac can directly induce cytoskeleton changes and affect gene transcription through activation of JNK.[31][32][33]
The noncanonical Wnt/Ca2+pathway[edit]

The noncanonical Wnt/Ca2+pathway regulates intracellularcalciumlevels. Again Wnt binds and activates to Frizzled. In this case however activated Frizzled causes a coupled G-protein to activate aphospholipase(PLC), which interacts with and splits PIP2into DAG and IP3.IP3can then bind to a receptor on theendoplasmic reticulumto release intracellular calcium stores, to induce calcium-dependent gene expression.[31][32][33]
Wnt signaling pathways and cancer[edit]
The Wnt signaling pathways are critical in cell-cell signaling during normal development and embryogenesis and required for maintenance of adult tissue, therefore it is not difficult to understand why disruption in Wnt signaling pathways can promote humandegenerative diseaseandcancer.
The Wnt signaling pathways are complex, involving many different elements, and therefore have many targets for misregulation. Mutations that cause constitutive activation of the Wnt signaling pathway lead to tumor formation and cancer. Aberrant activation of the Wnt pathway can lead to increase cell proliferation. Current research is focused on the action of the Wnt signaling pathway the regulation of stem cell choice to proliferate and self renew. This action of Wnt signaling in the possible control and maintenance of stem cells, may provide a possible treatment in cancers exhibiting aberrant Wnt signaling.[34][35][36]
TGF-β superfamily[edit]
"TGF"(Transforming Growth Factor) is a family of proteins that includes 33 members that encodedimeric,secreted polypeptides that regulate development.[37]Many developmental processes are under its control including gastrulation, axis symmetry of the body, organ morphogenesis, and tissue homeostasis in adults.[38]AllTGF-βligands bind to either Type I or Type II receptors, to create heterotetramic complexes.[39]
TGF-β pathway[edit]
TheTGF-β pathwayregulates many cellular processes in developing embryo and adult organisms, includingcell growth,differentiation,apoptosis,andhomeostasis.There are five kinds of type II receptors and seven types of type I receptors in humans and other mammals. These receptors are known as "dual-specificity kinases" because their cytoplasmic kinase domain has weak tyrosine kinase activity but strongserine/threoninekinase activity.[40]When a TGF-β superfamily ligand binds to the type II receptor, it recruits a type I receptor and activates it by phosphorylating the serine or threonine residues of its "GS" box.[41]This forms an activation complex that can then phosphorylate SMAD proteins.

SMAD pathway[edit]
There are three classes of SMADs:
Examples of SMADs in each class:[42][43][44]
| Class | SMADs |
|---|---|
| R-SMAD | SMAD1,SMAD2,SMAD3,SMAD5andSMAD8/9 |
| Co-SMAD | SMAD4 |
| I-SMAD | SMAD6andSMAD7 |
The TGF-β superfamily activates members of theSMADfamily, which function as transcription factors. Specifically, the type I receptor, activated by the type II receptor, phosphorylatesR-SMADsthat then bind to the co-SMAD,SMAD4.The R-SMAD/Co-SMAD forms a complex withimportinand enters the nucleus, where they act astranscription factorsand either up-regulate or down-regulate in the expression of a target gene.
Specific TGF-β ligands will result in the activation of either the SMAD2/3 or the SMAD1/5R-SMADs.For instance, whenactivin,Nodal,orTGF-β ligandbinds to the receptors, thephosphorylatedreceptor complex can activateSMAD2andSMAD3through phosphorylation. However, when a BMP ligand binds to the receptors, the phosphorylated receptor complex activatesSMAD1andSMAD5.Then, the Smad2/3 or the Smad1/5 complexes form a dimer complex withSMAD4and becometranscription factors.Though there are manyR-SMADsinvolved in the pathway, there is only one co-SMAD,SMAD4.[45]
Non-SMAD pathway[edit]
Non-Smad signaling proteins contribute to the responses of the TGF-β pathway in three ways. First, non-Smad signaling pathways phosphorylate the Smads. Second, Smads directly signal to other pathways by communicating directly with other signaling proteins, such as kinases. Finally, the TGF-β receptors directly phosphorylate non-Smad proteins.[46]
Members of TGF-β superfamily[edit]
1. TGF-β family[edit]
This family includesTGF-β1,TGF-β2,TGF-β3,and TGF-β5. They are involved in positively and negatively regulation ofcell division,the formation of theextracellular matrixbetween cells,apoptosis,andembryogenesis.They bind toTGF-β type II receptor(TGFBRII).
TGF-β1 stimulates the synthesis ofcollagenandfibronectinand inhibits the degradation of theextracellular matrix.Ultimately, it increases the production of extracellular matrix byepithelial cells.[39] TGF-β proteins regulate epithelia by controlling where and when they branch to form kidney, lung, and salivary gland ducts.[39]
2. Bone morphogenetic protein (BMPs) family[edit]
Members of the BMP family were originally found to inducebone formation,as their name suggests. However, BMPs are very multifunctional and can also regulateapoptosis,cell migration,cell division,anddifferentiation.They also specify the anterior/posterior axis, induce growth, and regulatehomeostasis.[37]
The BMPs bind to thebone morphogenetic protein receptor type II(BMPR2). Some of the proteins of theBMPfamily areBMP4andBMP7.BMP4promotes bone formation, causes cell death, or signals the formation ofepidermis,depending on the tissue it is acting on.BMP7is crucial for kidney development, sperm synthesis, and neural tube polarization. BothBMP4andBMP7regulate mature ligand stability and processing, including degrading ligands in lysosomes.[37]BMPs act by diffusing from the cells that create them.[47]
Other members of TGF-β superfamily[edit]
- Vg1 Family
- Activin Family
- Involved inembryogenesisandosteogenesis
- Regulateinsulinandpituitary,gonadal, andhypothalamichormones
- Nerve cell survival factors
- 3 Activins:Activin A,Activin BandActivin AB.
- Glial-Derived Neurotrophic Factor (GDNF)
- Needed for kidney andenteric neurondifferentiation
- Müllerian Inhibitory Factor
- Involved in mammalian sex determination
- Nodal
- Binds toActivin A Type 2B receptor
- Forms receptor complex withActivin A Type 1B receptoror withActivin A Type 1C receptor.[48]
- Growth and differentiation factors (GDFs)
Summary table of TGF-β signaling pathway[edit]
| TGF Beta superfamily ligand | Type II Receptor | Type I Receptor | R-SMADs | Co-SMAD | Ligand Inhibitors |
|---|---|---|---|---|---|
| Activin A | ACVR2A | ACVR1B(ALK4) | SMAD2,SMAD3 | SMAD4 | Follistatin |
| GDF1 | ACVR2A | ACVR1B(ALK4) | SMAD2,SMAD3 | SMAD4 | |
| GDF11 | ACVR2B | ACVR1B(ALK4),TGFβRI(ALK5) | SMAD2,SMAD3 | SMAD4 | |
| Bone morphogenetic proteins | BMPR2 | BMPR1A(ALK3),BMPR1B(ALK6) | SMAD1SMAD5,SMAD8 | SMAD4 | Noggin,Chordin,DAN |
| Nodal | ACVR2B | ACVR1B(ALK4),ACVR1C(ALK7) | SMAD2,SMAD3 | SMAD4 | Lefty |
| TGFβs | TGFβRII | TGFβRI(ALK5) | SMAD2,SMAD3 | SMAD4 | LTBP1,THBS1,Decorin |
Examples[edit]
Growth factorandclotting factorsare paracrine signaling agents. The local action of growth factor signaling plays an especially important role in the development of tissues. Also,retinoic acid,the active form ofvitamin A,functions in a paracrine fashion to regulate gene expression during embryonic development in higher animals.[49] In insects,Allatostatincontrols growth through paracrine action on the corpora allata.[citation needed]
In mature organisms, paracrine signaling is involved in responses toallergens,tissue repair, the formation ofscar tissue,and bloodclotting.[citation needed]Histamineis a paracrine that is released by immune cells in the bronchial tree. Histamine causes the smooth muscle cells of the bronchi to constrict, narrowing the airways.[50]
See also[edit]
- cAMP dependent pathway
- Crosstalk (biology)
- Lipid signaling
- Local hormone– either a paracrine hormone, or a hormone acting in both a paracrine and an endocrine fashion
- MAPK signaling pathway
- Netpath– A curated resource of signal transduction pathways in humans
- Paracrine regulator
References[edit]
- ^"Paracrine Factors".Retrieved27 July2018.
- ^Gospodarowicz, D.; Ferrara, N.; Schweigerer, L.; Neufeld, G. (1987). "Structural Characterization and Biological Functions of Fibroblast Growth Factor".Endocrine Reviews.8(2): 95–114.doi:10.1210/edrv-8-2-95.PMID2440668.
- ^Rifkin, Daniel B.; Moscatelli, David (1989)."Recent developments in the cell biology of basic fibroblast growth factor".The Journal of Cell Biology.109(1): 1–6.doi:10.1083/jcb.109.1.1.JSTOR1613457.PMC2115467.PMID2545723.
- ^abLappi, Douglas A. (1995). "Tumor targeting through fibroblast growth factor receptors".Seminars in Cancer Biology.6(5): 279–88.doi:10.1006/scbi.1995.0036.PMID8562905.
- ^abXu, J.; Xu, J; Colvin, JS; McEwen, DG; MacArthur, CA; Coulier, F; Gao, G; Goldfarb, M (1996)."Receptor Specificity of the Fibroblast Growth Factor Family".Journal of Biological Chemistry.271(25): 15292–7.doi:10.1074/jbc.271.25.15292.PMID8663044.
- ^Logan, M. (2003)."Finger or toe: The molecular basis of limb identity".Development.130(26): 6401–10.doi:10.1242/dev.00956.PMID14660539.
- ^abFantl, Wendy J; Johnson, Daniel E; Williams, Lewis T (1993). "Signaling by Receptor Tyrosine Kinases".Annual Review of Biochemistry.62:453–81.doi:10.1146/annurev.bi.62.070193.002321.PMID7688944.
- ^Yarden, Yosef; Ullrich, Axel (1988). "Growth Factor Receptor Tyrosine Kinases".Annual Review of Biochemistry.57:443–78.doi:10.1146/annurev.bi.57.070188.002303.PMID3052279.
- ^Katz, Michael E; McCormick, Frank (1997). "Signal transduction from multiple Ras effectors".Current Opinion in Genetics & Development.7(1): 75–9.doi:10.1016/S0959-437X(97)80112-8.PMID9024640.
- ^Zsebo, Krisztina M.; Williams, David A.; Geissler, Edwin N.; Broudy, Virginia C.; Martin, Francis H.; Atkins, Harry L.; Hsu, Rou-Yin; Birkett, Neal C.; Okino, Kenneth H.; Murdock, Douglas C.; Jacobsen, Frederick W.; Langley, Keith E.; Smith, Kent A.; Takeish, Takashi; Cattanach, Bruce M.; Galli, Stephen J.; Suggs, Sidney V. (1990). "Stem cell factor is encoded at the SI locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor".Cell.63(1): 213–24.doi:10.1016/0092-8674(90)90302-U.PMID1698556.S2CID39924379.
- ^Rönnstrand, L. (2004). "Signal transduction via the stem cell factor receptor/c-Kit".Cellular and Molecular Life Sciences.61(19–20): 2535–48.doi:10.1007/s00018-004-4189-6.PMID15526160.S2CID2602233.
- ^Melillo, Rosa Marina; Castellone, Maria Domenica; Guarino, Valentina; De Falco, Valentina; Cirafici, Anna Maria; Salvatore, Giuliana; Caiazzo, Fiorina; Basolo, Fulvio; Giannini, Riccardo; Kruhoffer, Mogens; Orntoft, Torben; Fusco, Alfredo; Santoro, Massimo (2005)."The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells".Journal of Clinical Investigation.115(4): 1068–81.doi:10.1172/JCI22758.PMC1062891.PMID15761501.(Retracted, seedoi:10.1172/JCI87345,PMID27035814,Retraction Watch)
- ^Kolch, Walter (2000)."Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions".The Biochemical Journal.351(2): 289–305.doi:10.1042/0264-6021:3510289.PMC1221363.PMID11023813.
- ^abAaronson, David S.; Horvath, Curt M. (2002). "A Road Map for Those Who Don't Know JAK-STAT".Science.296(5573): 1653–5.Bibcode:2002Sci...296.1653A.doi:10.1126/science.1071545.PMID12040185.S2CID20857536.
- ^Rawlings, Jason S.; Rosler, Kristin M.; Harrison, Douglas A. (2004)."The JAK/STAT signaling pathway".Journal of Cell Science.117(8): 1281–3.doi:10.1242/jcs.00963.PMID15020666.
- ^O'Shea, John J; Gadina, Massimo; Schreiber, Robert D (2002)."Cytokine signaling in 2002: new surprises in the Jak/Stat pathway".Cell.109(2): S121–31.doi:10.1016/S0092-8674(02)00701-8.PMID11983158.
- ^Shiang, Rita; Thompson, Leslie M.; Zhu, Ya-Zhen; Church, Deanna M.; Fielder, Thomas J.; Bocian, Maureen; Winokur, Sara T.; Wasmuth, John J. (1994). "Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia".Cell.78(2): 335–42.doi:10.1016/0092-8674(94)90302-6.PMID7913883.S2CID20325070.
- ^Kalluri, Raghu; Weinberg, Robert A. (2009)."The basics of epithelial-mesenchymal transition".Journal of Clinical Investigation.119(6): 1420–8.doi:10.1172/JCI39104.PMC2689101.PMID19487818.
- ^Silver, Debra L.;Montell, Denise J.(2001)."Paracrine Signaling through the JAK/STAT Pathway Activates Invasive Behavior of Ovarian Epithelial Cells in Drosophila".Cell.107(7): 831–41.doi:10.1016/S0092-8674(01)00607-9.PMID11779460.
- ^Ingham, P. W.; McMahon, AP (2001)."Hedgehog signaling in animal development: Paradigms and principles".Genes & Development.15(23): 3059–87.doi:10.1101/gad.938601.PMID11731473.
- ^Bitgood, Mark J.; McMahon, Andrew P. (1995)."HedgehogandBmpGenes Are Coexpressed at Many Diverse Sites of Cell–Cell Interaction in the Mouse Embryo ".Developmental Biology.172(1): 126–38.doi:10.1006/dbio.1995.0010.PMID7589793.
- ^abJacob, L.; Lum, L. (2007). "Hedgehog Signaling Pathway".Science's STKE.2007(407): cm6.doi:10.1126/stke.4072007cm6.PMID17925577.S2CID35653781.
- ^Johnson, Ronald L; Scott, Matthew P (1998)."New players and puzzles in the Hedgehog signaling pathway".Current Opinion in Genetics & Development.8(4): 450–6.doi:10.1016/S0959-437X(98)80117-2.PMID9729722.
- ^Nybakken, K; Perrimon, N (2002). "Hedgehog signal transduction: Recent findings".Current Opinion in Genetics & Development.12(5): 503–11.doi:10.1016/S0959-437X(02)00333-7.PMID12200154.
- ^Lubik AA, Nouri M, Truong S, Ghaffari M, Adomat HH, Corey E, Cox ME, Li N, Guns ES, Yenki P, Pham S, Buttyan R (2016)."Paracrine Sonic Hedgehog Signaling Contributes Significantly to Acquired Steroidogenesis in the Prostate Tumor Microenvironment".International Journal of Cancer.140(2): 358–369.doi:10.1002/ijc.30450.PMID27672740.S2CID2354209.
- ^Collins, R. T.; Cohen, SM (2005)."A Genetic Screen in Drosophila for Identifying Novel Components of the Hedgehog Signaling Pathway".Genetics.170(1): 173–84.doi:10.1534/genetics.104.039420.PMC1449730.PMID15744048.
- ^Evangelista, M.; Tian, H.; De Sauvage, F. J. (2006)."The Hedgehog Signaling Pathway in Cancer".Clinical Cancer Research.12(20): 5924–8.doi:10.1158/1078-0432.CCR-06-1736.PMID17062662.
- ^Taipale, Jussi; Beachy, Philip A. (2001). "The Hedgehog and Wnt signaling pathways in cancer".Nature.411(6835): 349–54.Bibcode:2001Natur.411..349T.doi:10.1038/35077219.PMID11357142.S2CID4414768.
- ^Cadigan, K. M.; Nusse, R. (1997)."Wnt signaling: A common theme in animal development".Genes & Development.11(24): 3286–305.doi:10.1101/gad.11.24.3286.PMID9407023.
- ^abcDale, Trevor C. (1998)."Signal transduction by the Wnt family of ligands".The Biochemical Journal.329(Pt 2): 209–23.doi:10.1042/bj3290209.PMC1219034.PMID9425102.
- ^abcChen, Xi; Yang, Jun; Evans, Paul M; Liu, Chunming (2008)."Wnt signaling: The good and the bad".Acta Biochimica et Biophysica Sinica.40(7): 577–94.doi:10.1111/j.1745-7270.2008.00440.x.PMC2532600.PMID18604449.
- ^abcKomiya, Yuko; Habas, Raymond (2008)."Wnt signal transduction pathways".Organogenesis.4(2): 68–75.doi:10.4161/org.4.2.5851.PMC2634250.PMID19279717.
- ^Logan, Catriona Y.; Nusse, Roel (2004). "The Wnt Signaling Pathway in Development and Disease".Annual Review of Cell and Developmental Biology.20:781–810.CiteSeerX10.1.1.322.311.doi:10.1146/annurev.cellbio.20.010403.113126.PMID15473860.
- ^Lustig, B; Behrens, J (2003). "The Wnt signaling pathway and its role in tumor development".Journal of Cancer Research and Clinical Oncology.129(4): 199–221.doi:10.1007/s00432-003-0431-0.PMID12707770.S2CID28959851.
- ^Neth, Peter; Ries, Christian; Karow, Marisa; Egea, Virginia; Ilmer, Matthias; Jochum, Marianne (2007). "The Wnt Signal Transduction Pathway in Stem Cells and Cancer Cells: Influence on Cellular Invasion".Stem Cell Reviews.3(1): 18–29.doi:10.1007/s12015-007-0001-y.PMID17873378.S2CID25793825.
- ^abcBandyopadhyay, Amitabha; Tsuji, Kunikazu; Cox, Karen; Harfe, Brian D.; Rosen, Vicki; Tabin, Clifford J. (2006)."Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis".PLOS Genetics.2(12): e216.doi:10.1371/journal.pgen.0020216.PMC1713256.PMID17194222.
- ^Attisano, Liliana; Wrana, Jeffrey L. (2002). "Signal Transduction by the TGF-β Superfamily".Science.296(5573): 1646–7.Bibcode:2002Sci...296.1646A.doi:10.1126/science.1071809.PMID12040180.S2CID84138159.
- ^abcWrana, Jeffrey L.; Ozdamar, Barish; Le Roy, Christine; Benchabane, Hassina (2008)."Signaling Receptors of the TGF-β Family".In Derynck, Rik; Miyazono, Kohei (eds.).The TGF-β Family.CSHL Press. pp. 151–77.ISBN978-0-87969-752-5.
- ^ten Dijke, Peter; Heldin, Carl-Henrik (2006)."The Smad family".In ten Dijke, Peter; Heldin, Carl-Henrik (eds.).Smad Signal Transduction: Smads in Proliferation, Differentiation and Disease.Proteins and Cell Regulation. Vol. 5. Dordrecht: Springer. pp. 1–13.ISBN978-1-4020-4709-1.
- ^Moustakas, Aristidis (2002-09-01)."Smad signaling network".Journal of Cell Science.115(17): 3355–6.doi:10.1242/jcs.115.17.3355.PMID12154066.
- ^Wu, Jia-Wei; Hu, Min; Chai, Jijie; Seoane, Joan; Huse, Morgan; Li, Carey; Rigotti, Daniel J.; Kyin, Saw; Muir, Tom W.; Fairman, Robert; Massagué, Joan; Shi, Yigong (2001)."Crystal Structure of a Phosphorylated Smad2".Molecular Cell.8(6): 1277–89.doi:10.1016/S1097-2765(01)00421-X.PMID11779503.
- ^Pavletich, Nikola P.; Hata, Yigong; Lo, Akiko; Massagué, Roger S.; Pavletich, Joan (1997)."A structural basis for mutational inactivation of the tumour suppressor Smad4".Nature.388(6637): 87–93.Bibcode:1997Natur.388R..87S.doi:10.1038/40431.PMID9214508.
- ^Itoh, Fumiko; Asao, Hironobu; Sugamura, Kazuo; Heldin, Carl-Henrik; Ten Dijke, Peter; Itoh, Susumu (2001)."Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads".The EMBO Journal.20(15): 4132–42.doi:10.1093/emboj/20.15.4132.PMC149146.PMID11483516.
- ^Schmierer, Bernhard; Hill, Caroline S. (2007). "TGFβ–SMAD signal transduction: Molecular specificity and functional flexibility".Nature Reviews Molecular Cell Biology.8(12): 970–82.doi:10.1038/nrm2297.PMID18000526.S2CID131895.
- ^Moustakas, Aristidis; Heldin, Carl-Henrik (2005)."Non-Smad TGF-β signals".Journal of Cell Science.118(16): 3573–84.doi:10.1242/jcs.02554.PMID16105881.
- ^Ohkawara, Bisei; Iemura, Shun-Ichiro; Ten Dijke, Peter; Ueno, Naoto (2002)."Action Range of BMP is Defined by Its N-Terminal Basic Amino Acid Core".Current Biology.12(3): 205–9.doi:10.1016/S0960-9822(01)00684-4.PMID11839272.
- ^Munir, Sadia; Xu, Guoxiong; Wu, Yaojiong; Yang, Burton; Lala, Peeyush K.; Peng, Chun (2004)."Nodal and ALK7 Inhibit Proliferation and Induce Apoptosis in Human Trophoblast Cells".Journal of Biological Chemistry.279(30): 31277–86.doi:10.1074/jbc.M400641200.PMID15150278.
- ^Duester, Gregg (September 2008)."Retinoic acid synthesis and signaling during early organogenesis".Cell.134(6): 921–31.doi:10.1016/j.cell.2008.09.002.PMC2632951.PMID18805086.
- ^
 This article incorporatestextavailable under theCC BY 4.0license.Betts, J Gordon; Desaix, Peter; Johnson, Eddie; Johnson, Jody E; Korol, Oksana; Kruse, Dean; Poe, Brandon; Wise, James; Womble, Mark D; Young, Kelly A (July 24, 2023).Anatomy & Physiology.Houston: OpenStax CNX. 17.1 Overview of the endocrine system.ISBN978-1-947172-04-3.
This article incorporatestextavailable under theCC BY 4.0license.Betts, J Gordon; Desaix, Peter; Johnson, Eddie; Johnson, Jody E; Korol, Oksana; Kruse, Dean; Poe, Brandon; Wise, James; Womble, Mark D; Young, Kelly A (July 24, 2023).Anatomy & Physiology.Houston: OpenStax CNX. 17.1 Overview of the endocrine system.ISBN978-1-947172-04-3.
External links[edit]
- Paracrine+Signalingat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
- "paracrine"atDorland's Medical Dictionary
