Proteome

Theproteomeis the entire set ofproteinsthat is, or can be, expressed by agenome,cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions.Proteomicsis the study of the proteome.
Types of proteomes[edit]
While proteome generally refers to the proteome of an organism, multicellular organisms may have very different proteomes in different cells, hence it is important to distinguish proteomes in cells and organisms.
Acellular proteomeis the collection of proteins found in a particularcelltype under a particular set of environmental conditions such as exposure tohormone stimulation.
It can also be useful to consider an organism'scomplete proteome,which can be conceptualized as the complete set of proteins from all of the various cellular proteomes. This is very roughly the protein equivalent of thegenome.
The termproteomehas also been used to refer to the collection of proteins in certainsub-cellular systems,such as organelles. For instance, themitochondrialproteome may consist of more than 3000 distinct proteins.[1][2][3]
The proteins in aviruscan be called aviral proteome.Usually viral proteomes are predicted from the viral genome[4]but some attempts have been made to determine all the proteins expressed from a virus genome, i.e. the viral proteome.[5]More often, however, virus proteomics analyzes the changes of host proteins upon virus infection, so that in effecttwoproteomes (of virus and its host) are studied.[6]
Importance in cancer[edit]
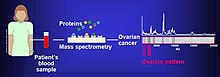
The proteome can be used in order to comparatively analyze different cancer cell lines. Proteomic studies have been used in order to identify the likelihood of metastasis in bladder cancer cell lines KK47 and YTS1 and were found to have 36 unregulated and 74 down regulated proteins.[7]The differences in protein expression can help identify novel cancer signaling mechanisms.
Biomarkersof cancer have been found bymass spectrometrybased proteomic analyses. The use of proteomics or the study of the proteome is a step forward in personalized medicine to tailor drug cocktails to the patient's specific proteomic and genomic profile.[8]The analysis of ovarian cancer cell lines showed that putative biomarkers for ovarian cancer include "α-enolase (ENOA),elongation factor Tu,mitochondrial (EFTU),glyceraldehyde-3-phosphate dehydrogenase (G3P),stress-70 protein, mitochondrial (GRP75),apolipoprotein A-1 (APOA1),peroxiredoxin (PRDX2) andannexin A (ANXA)".[9]
Comparative proteomic analyses of 11 cell lines demonstrated the similarity between the metabolic processes of each cell line; 11,731 proteins were completely identified from this study. Housekeeping proteins tend to show greater variability between cell lines.[10]
Resistance to certain cancer drugs is still not well understood. Proteomic analysis has been used in order to identify proteins that may have anti-cancer drug properties, specifically for the colon cancer drugirinotecan.[11]Studies of adenocarcinoma cell line LoVo demonstrated that 8 proteins were unregulated and 7 proteins were down-regulated. Proteins that showed a differential expression were involved in processes such as transcription,apoptosisand cell proliferation/differentiation among others.
The proteome in bacterial systems[edit]
Proteomic analyses have been performed in different kinds of bacteria to assess their metabolic reactions to different conditions. For example, in bacteria such asClostridiumandBacillus,proteomic analyses were used in order to investigate how different proteins help each of these bacteria spores germinate after a prolonged period of dormancy.[12]In order to better understand how to properly eliminate spores, proteomic analysis must be performed.
History[edit]
Marc Wilkinscoined the termproteome[13]in 1994 in a symposium on "2D Electrophoresis: from protein maps to genomes" held in Siena in Italy. It appeared in print in 1995,[14]with the publication of part of his PhD thesis. Wilkins used the term to describe the entire complement ofproteinsexpressed by a genome, cell, tissue or organism.
Size and contents[edit]
The genomes ofvirusesandprokaryotesencode a relatively well-defined proteome as each protein can be predicted with high confidence, based on itsopen reading frame(in viruses ranging from ~3 to ~1000, in bacteria ranging from about 500 proteins to about 10,000).[15]However, mostprotein predictionalgorithms use certain cut-offs, such as 50 or 100 amino acids, so small proteins are often missed by such predictions.[16]Ineukaryotesthis becomes much more complicated as more than oneproteincan be produced from mostgenesdue toalternative splicing(e.g. human proteome encodes about 20,000 proteins, but some estimates predicted 92,179 proteins[citation needed]out of which 71,173 are splicing variants[citation needed]).[17]
Association of proteome size with DNA repair capability
The concept of “proteomic constraint” is thatDNA repaircapacity is positively correlated with the information content of agenome,which, in turn, is approximately related to the size of the proteome.[18]Inbacteria,archaeaandDNA viruses,DNA repair capability is positively related to genome information content and to genome size.[18]“Proteomic constraint” proposes that modulators of mutation rates such as DNA repair genes are subject to selection pressure proportional to the amount of information in a genome.[18]
Proteoforms.There are different factors that can add variability to proteins. SAPs (single amino acid polymorphisms) and non-synonymous single nucleotide polymorphisms (nsSNPs) can lead to different "proteoforms"[19]or "proteomorphs". Recent estimates have found ~135,000 validated nonsynonymous cSNPs currently housed within SwissProt. In dbSNP, there are 4.7 million candidate cSNPs, yet only ~670,000 cSNPs have been validated in the 1,000-genomes set as nonsynonymous cSNPs that change the identity of an amino acid in a protein.[19]
Dark proteome.The termdark proteomecoined by Perdigão and colleagues, defines regions of proteins that have no detectablesequence homologyto other proteins of knownthree-dimensional structureand therefore cannot bemodeled by homology.For 546,000 Swiss-Prot proteins, 44–54% of the proteome ineukaryotesand viruses was found to be "dark", compared with only ~14% inarchaeaandbacteria.[20]
Human proteome.Currently, several projects aim to map the human proteome, including theHuman Proteome Map,ProteomicsDB,isoform.io,andThe Human Proteome Project (HPP).Much like thehuman genome project,these projects seek to find and collect evidence for all predicted protein coding genes in the human genome. The Human Proteome Map currently (October 2020) claims 17,294 proteins and ProteomicsDB 15,479, using different criteria. On October 16, 2020, the HPP published a high-stringency blueprint[21]covering more than 90% of the predicted protein coding genes. Proteins are identified from a wide range of fetal and adult tissues and cell types, includinghematopoietic cells.
Methods to study the proteome[edit]

Analyzing proteins proves to be more difficult than analyzing nucleic acid sequences. While there are only 4 nucleotides that make up DNA, there are at least 20 different amino acids that can make up a protein. Additionally, there is currently no knownhigh throughputtechnology to make copies of a single protein. Numerous methods are available to study proteins, sets of proteins, or the whole proteome. In fact, proteins are often studied indirectly, e.g. using computational methods and analyses of genomes. Only a few examples are given below.
Separation techniques and electrophoresis[edit]
Proteomics,the study of the proteome, has largely been practiced through the separation of proteins bytwo dimensional gel electrophoresis.In the first dimension, the proteins are separated byisoelectric focusing,which resolves proteins on the basis of charge. In the second dimension, proteins are separated bymolecular weightusingSDS-PAGE.The gel isstainedwithCoomassie brilliant blueorsilverto visualize the proteins. Spots on the gel are proteins that have migrated to specific locations.
Mass spectrometry[edit]

Mass spectrometryis one of the key methods to study the proteome.[22]Some important mass spectrometry methods include Orbitrap Mass Spectrometry,MALDI(Matrix Assisted Laser Desorption/Ionization), andESI (Electrospray Ionization).Peptide mass fingerprintingidentifies a protein by cleaving it into short peptides and then deduces the protein's identity by matching the observed peptide masses against asequence database.Tandem mass spectrometry,on the other hand, can get sequence information from individual peptides by isolating them, colliding them with a non-reactive gas, and then cataloguing the fragmentionsproduced.[23]
In May 2014, a draft map of the human proteome was published inNature.[24]This map was generated using high-resolution Fourier-transform mass spectrometry. This study profiled 30 histologically normal human samples resulting in the identification of proteins coded by 17,294 genes. This accounts for around 84% of the total annotated protein-coding genes.
Chromatography[edit]
Liquidchromatographyis an important tool in the study of the proteome. It allows for very sensitive separation of different kinds of proteins based on their affinity for a matrix. Some newer methods for the separation and identification of proteins include the use of monolithic capillary columns, high temperature chromatography and capillary electrochromatography.[25]
Blotting[edit]
Western blottingcan be used in order to quantify the abundance of certain proteins. By using antibodies specific to the protein of interest, it is possible to probe for the presence of specific proteins from a mixture of proteins.
Protein complementation assays and interaction screens[edit]
Protein-fragment complementation assaysare often used to detectprotein–protein interactions.Theyeast two-hybrid assayis the most popular of them but there are numerous variations, both usedin vitroandin vivo.Pull-down assays are a method to determine the protein binding partners of a given protein.[26]
Protein structure prediction[edit]
Protein structure predictioncan be used to provide three-dimensional protein structure predictions of whole proteomes. In 2022, a large-scale collaboration betweenEMBL-EBIandDeepMindprovided predicted structures for over 200 million proteins from across the tree of life.[27]Smaller projects have also used protein structure prediction to help map the proteome of individual organisms, for exampleisoform.ioprovides coverage of multiple protein isoforms for over 20,000 genes in thehuman genome.[28]
Protein databases[edit]
TheHuman Protein Atlascontains information about the human proteins in cells, tissues, and organs. All the data in the knowledge resource is open access to allow scientists both in academia and industry to freely access the data for exploration of the human proteome. The organizationELIXIRhas selected the protein atlas as a core resource due to its fundamental importance for a wider life science community.
ThePlasma Proteome databasecontains information on 10,500blood plasmaproteins. Because the range in protein contents in plasma is very large, it is difficult to detect proteins that tend to be scarce when compared to abundant proteins. This is an analytical limit that may possibly be a barrier for the detections of proteins with ultra low concentrations.[29]
Databases such asneXtprotandUniProtare central resources for human proteomic data.
See also[edit]
- Metabolome
- Cytome
- Bioinformatics
- List of omics topics in biology
- Plant Proteome Database
- Transcriptome
- Interactome
- Human Proteome Project
- BioPlex
- Human Protein Atlas
References[edit]
- ^Johnson, D. T.; Harris, R. A.; French, S.; Blair, P. V.; You, J.; Bemis, K. G.; Wang, M.; Balaban, R. S. (2006)."Tissue heterogeneity of the mammalian mitochondrial proteome".American Journal of Physiology. Cell Physiology.292(2): c689–c697.doi:10.1152/ajpcell.00108.2006.PMID16928776.S2CID24412700.
- ^Morgenstern, Marcel; Stiller, Sebastian B.; Lübbert, Philipp; Peikert, Christian D.; Dannenmaier, Stefan; Drepper, Friedel; Weill, Uri; Höß, Philipp; Feuerstein, Reinhild; Gebert, Michael; Bohnert, Maria (June 2017)."Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale".Cell Reports.19(13): 2836–2852.doi:10.1016/j.celrep.2017.06.014.ISSN2211-1247.PMC5494306.PMID28658629.
- ^Gómez-Serrano, M (November 2018)."Mitoproteomics: Tackling Mitochondrial Dysfunction in Human Disease".Oxid Med Cell Longev.2018:1435934.doi:10.1155/2018/1435934.PMC6250043.PMID30533169.
- ^Uetz, P. (2004-10-15)."From ORFeomes to Protein Interaction Maps in Viruses".Genome Research.14(10b): 2029–2033.doi:10.1101/gr.2583304.ISSN1088-9051.PMID15489322.
- ^Maxwell, Karen L.; Frappier, Lori (June 2007)."Viral proteomics".Microbiology and Molecular Biology Reviews.71(2): 398–411.doi:10.1128/MMBR.00042-06.ISSN1092-2172.PMC1899879.PMID17554050.
- ^Viswanathan, Kasinath; Früh, Klaus (December 2007)."Viral proteomics: global evaluation of viruses and their interaction with the host".Expert Review of Proteomics.4(6): 815–829.doi:10.1586/14789450.4.6.815.ISSN1744-8387.PMID18067418.S2CID25742649.
- ^Yang, Ganglong; Xu, Zhipeng; Lu, Wei; Li, Xiang; Sun, Chengwen; Guo, Jia; Xue, Peng; Guan, Feng (2015-07-31)."Quantitative Analysis of Differential Proteome Expression in Bladder Cancer vs. Normal Bladder Cells Using SILAC Method".PLOS ONE.10(7): e0134727.Bibcode:2015PLoSO..1034727Y.doi:10.1371/journal.pone.0134727.ISSN1932-6203.PMC4521931.PMID26230496.
- ^An, Yao; Zhou, Li; Huang, Zhao; Nice, Edouard C.; Zhang, Haiyuan; Huang, Canhua (2019-05-04). "Molecular insights into cancer drug resistance from a proteomics perspective".Expert Review of Proteomics.16(5): 413–429.doi:10.1080/14789450.2019.1601561.ISSN1478-9450.PMID30925852.S2CID88474614.
- ^Cruz, Isa N.; Coley, Helen M.; Kramer, Holger B.; Madhuri, Thumuluru Kavitah; Safuwan, Nur a. M.; Angelino, Ana Rita; Yang, Min (2017-01-01)."Proteomics Analysis of Ovarian Cancer Cell Lines and Tissues Reveals Drug Resistance-associated Proteins".Cancer Genomics & Proteomics.14(1): 35–51.doi:10.21873/cgp.20017.ISSN1109-6535.PMC5267499.PMID28031236.
- ^Geiger, Tamar; Wehner, Anja; Schaab, Christoph; Cox, Juergen; Mann, Matthias (March 2012)."Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins".Molecular & Cellular Proteomics.11(3): M111.014050.doi:10.1074/mcp.M111.014050.ISSN1535-9476.PMC3316730.PMID22278370.
- ^Peng, Xing-Chen; Gong, Feng-Ming; Wei, Meng; Chen, Xi; Chen, Ye; Cheng, Ke; Gao, Feng; Xu, Feng; Bi, Feng; Liu, Ji-Yan (December 2010). "Proteomic analysis of cell lines to identify the irinotecan resistance proteins".Journal of Biosciences.35(4): 557–564.doi:10.1007/s12038-010-0064-9.ISSN0250-5991.PMID21289438.S2CID6082637.
- ^Chen, Yan; Barat, Bidisha; Ray, W. Keith; Helm, Richard F.; Melville, Stephen B.; Popham, David L. (2019-03-15)."Membrane Proteomes and Ion Transporters in Bacillus anthracis and Bacillus subtilis Dormant and Germinating Spores".Journal of Bacteriology.201(6).doi:10.1128/JB.00662-18.ISSN0021-9193.PMC6398275.PMID30602489.
- ^Wilkins, Marc (Dec 2009). "Proteomics data mining".Expert Review of Proteomics.6(6).England:599–603.doi:10.1586/epr.09.81.PMID19929606.S2CID207211912.
- ^Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I (1995). "Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium".Electrophoresis.16(1): 1090–94.doi:10.1002/elps.11501601185.PMID7498152.S2CID9269742.
- ^Kozlowski, LP (26 October 2016)."Proteome-pI:proteome isoelectric point database ".Nucleic Acids Research.45(D1): D1112–D1116.doi:10.1093/nar/gkw978.PMC5210655.PMID27789699.
- ^Leslie, Mitch (2019-10-18)."Outsize impact".Science.366(6463): 296–299.Bibcode:2019Sci...366..296L.doi:10.1126/science.366.6463.296.ISSN0036-8075.PMID31624194.S2CID204774732.
- ^Uniprot, Consortium (2014)."UniProt: a hub for protein information".Nucleic Acids Research.43(D1): D204–D212.doi:10.1093/nar/gku989.ISSN0305-1048.PMC4384041.PMID25348405.
- ^abcAcosta S, Carela M, Garcia-Gonzalez A, Gines M, Vicens L, Cruet R, Massey SE. DNA Repair Is Associated with Information Content in Bacteria, Archaea, and DNA Viruses. J Hered. 2015 Sep-Oct;106(5):644-59. doi: 10.1093/jhered/esv055. Epub 2015 Aug 29. PMID: 26320243
- ^abAebersold, Ruedi; Agar, Jeffrey N; Amster, I Jonathan; Baker, Mark S; Bertozzi, Carolyn R; Boja, Emily S; Costello, Catherine E; Cravatt, Benjamin F; Fenselau, Catherine; Garcia, Benjamin A; Ge, Ying (March 2018)."How many human proteoforms are there?".Nature Chemical Biology.14(3): 206–214.doi:10.1038/nchembio.2576.hdl:1721.1/120977.ISSN1552-4450.PMC5837046.PMID29443976.
- ^Perdigão, Nelson; et al. (2015)."Unexpected features of the dark proteome".PNAS.112(52): 15898–15903.Bibcode:2015PNAS..11215898P.doi:10.1073/pnas.1508380112.PMC4702990.PMID26578815.
- ^Adhikari, S (October 2020)."A high-stringency blueprint of the human proteome".Nature Communications.11(1): 5301.Bibcode:2020NatCo..11.5301A.doi:10.1038/s41467-020-19045-9.PMC7568584.PMID33067450.
- ^Altelaar, AF; Munoz, J; Heck, AJ (January 2013). "Next-generation proteomics: towards an integrative view of proteome dynamics".Nature Reviews Genetics.14(1): 35–48.doi:10.1038/nrg3356.PMID23207911.S2CID10248311.
- ^Wilhelm, Mathias; Schlegl, Judith; Hahne, Hannes; Gholami, Amin Moghaddas; Lieberenz, Marcus; Savitski, Mikhail M.; Ziegler, Emanuel; Butzmann, Lars; Gessulat, Siegfried; Marx, Harald; Mathieson, Toby; Lemeer, Simone; Schnatbaum, Karsten; Reimer, Ulf; Wenschuh, Holger; Mollenhauer, Martin; Slotta-Huspenina, Julia; Boese, Joos-Hendrik; Bantscheff, Marcus; Gerstmair, Anja; Faerber, Franz; Kuster, Bernhard (2014)."Mass-Spectrometry-Based Draft of the Human Proteome".Nature.509(7502): 582–7.Bibcode:2014Natur.509..582W.doi:10.1038/nature13319.PMID24870543.S2CID4467721.
- ^Kim, Min-Sik; et al. (May 2014)."A draft map of the human proteome".Nature.509(7502): 575–81.Bibcode:2014Natur.509..575K.doi:10.1038/nature13302.PMC4403737.PMID24870542.
- ^Shi, Yang; Xiang, Rong; Horváth, Csaba; Wilkins, James A. (2004-10-22). "The role of liquid chromatography in proteomics".Journal of Chromatography A.Bioanalytical Chemistry: Perspectives and Recent Advances with Recognition of Barry L. Karger.1053(1): 27–36.doi:10.1016/j.chroma.2004.07.044.ISSN0021-9673.PMID15543969.
- ^"Pull-Down Assays - US".www.thermofisher.com.Retrieved2019-12-05.
- ^Callaway, Ewen (2022-07-28)."'The entire protein universe': AI predicts shape of nearly every known protein ".Nature.608(7921): 15–16.Bibcode:2022Natur.608...15C.doi:10.1038/d41586-022-02083-2.PMID35902752.S2CID251159714.
- ^Sommer, Markus J.; Cha, Sooyoung; Varabyou, Ales; Rincon, Natalia; Park, Sukhwan; Minkin, Ilia; Pertea, Mihaela; Steinegger, Martin; Salzberg, Steven L. (2022-12-15)."Structure-guided isoform identification for the human transcriptome".eLife.11:e82556.doi:10.7554/eLife.82556.PMC9812405.PMID36519529.
- ^Ponomarenko, Elena A.; Poverennaya, Ekaterina V.; Ilgisonis, Ekaterina V.; Pyatnitskiy, Mikhail A.; Kopylov, Arthur T.; Zgoda, Victor G.; Lisitsa, Andrey V.; Archakov, Alexander I. (2016)."The Size of the Human Proteome: The Width and Depth".International Journal of Analytical Chemistry.2016:7436849.doi:10.1155/2016/7436849.ISSN1687-8760.PMC4889822.PMID27298622.
External links[edit]
- PIR database
- UniProt database
- Pfam databaseat theLibrary of CongressWeb Archives (archived 2011-05-06)
