Smoothened
Smoothenedis aproteinthat in humans is encoded by theSMOgene.Smoothened is a ClassFrizzled(Class F)G protein-coupled receptor[5][6]that is a component of thehedgehog signaling pathwayand is conserved fromfliesto humans. It is the molecular target of the naturalteratogencyclopamine.[7]It also is the target ofvismodegib,the first hedgehog pathway inhibitor to be approved by the U.S.Food and Drug Administration (FDA).[8]
Smoothened (Smo) is a keytransmembrane proteinthat is a key component of thehedgehog signaling pathway,a cell-cell communication system critical for embryonic development and adult tissuehomeostasis.[9][10]Mutations in proteins that relay Hh signals between cells cause birth defects andcancer.[11]The protein that carries the Hh signal across the membrane is theoncoproteinandG-protein coupled receptor(GPCR) Smoothened (Smo). Smo is regulated by a separate transmembrane receptor for Hh ligands calledPatched(Ptc). Ptc itself is atumor suppressorthat keeps the Hh pathway off by inhibiting Smo. The excessive Hh signaling that drives human skin and brain cancer is most frequently caused by inactivating mutations in Ptc or by gain of function mutations in Smo. While direct Smoagonistsandantagonists,such as SAG andvismodegib,can bind to and activate or inhibit Smo, how Ptc inhibits Smo endogenously remains a mystery in the field.
Currently, Smo is targeted and inhibited directly by a small-molecule drug, vismodegib, for the treatment of advanced basal cell cancer, however widespread resistance to this drug has become a prevalent issue.[12][13]Finding another method to target Smo activity in Hh-driven cancers would provide valuable information for novel therapeutics. Identifying these Ptc responsive sites on Smo will help solve a long-standing mystery in Hh signaling and suggest new therapeutic strategies to block Smo activity in Hh-driven cancers.
Function[edit]
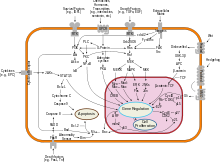
Cellular localization plays an essential role in the function of SMO, which anchors to the cell membrane as a 7-pass transmembrane protein. Stimulation of thepatched12-pass transmembrane receptor by thesonic hedgehogligand leads to translocation of SMO to theprimary ciliumin vertebrates in a process that involves the exit of patched from the primary cilium, where it normally localizes in its unstimulated state.[14]Vertebrate SMO that is mutated in the domain required for ciliary localisation often cannot contribute to hedgehog pathway activation.[15]Conversely, SMO can become constitutively localized to the primary cilium and potentially activate pathway signaling constitutively as a result of a tryptophan to leucine mutation in the aforementioned domain.[16]SMO has been shown to move during patched stimulation from the plasma membrane near the primary cilium to the ciliary membrane itself via a lateral transport pathway along the membrane, as opposed to via directed transport by vesicles. The cAMP-PKA pathway is known to promote the lateral movement of SMO and hedgehog signal transduction in general.[17]In invertebrates likeDrosophila,SMO does not organize at cilia and instead is generally translocated to the plasma membrane following hedgehog binding to patched.[18]
After cellular localization, SMO must additionally be activated by a distinct mechanism in order to stimulate hedgehog signal transduction, but that mechanism is unknown.[19]There is evidence for the existence of an unidentified endogenous ligand that binds SMO and activates it. It is believed that mutations in SMO can mimic the ligand-induced conformation of SMO and activate constitutive signal transduction.[18]
SMO plays a key role in transcriptional repression and activation by the zinc-finger transcription factorCubitus interruptus(Ci; known as Gli in vertebrates). When the hedgehog pathway is inactive, a complex of Fused (Fu), Suppressor of Fused (Sufu), and thekinesinmotor proteinCostal-2(Cos2) tether Ci to microtubules. In this complex, Cos2 promotes proteolytic cleavage of Ci by activatinghyperphosphorylationof Ci and subsequent recruitment of ubiquitin ligase; the cleaved Ci goes on to act as a repressor of hedgehog-activated transcription. However, when hedgehog signaling is active, Ci remains intact and acts as a transcriptional activator of the same genes that its cleaved form suppresses.[20][21]SMO has been shown to bind Costal-2 and play a role in the localization of the Ci complex and prevention of Ci cleavage.[22][23]Additionally, it is known that vertebrate SMO contributes to the activation of Gli as a transcription factor via association with ciliary structures such asEvc2,but these mechanisms are not fully understood.[18]
Endogenous activation[edit]

A leading hypothesis in the field is that Ptc regulates Smo by gating its access tocholesterolor a related sterol.[24]It has been proposed that cholesterol activates Smo, and subsequently Hh signaling, by entering the active site through a hydrophobic “oxysterol tunnel,” which can adopt open or closed conformations to allow for activation or inactivation of Smo, respectively, due to allowed sterol binding.[25][26]Shh would work by inhibiting Ptc, which would increase accessible cholesterol concentrations and allow for the activation of Smo and transmission of the Hh signal.[27] A recent crystal structure has identified two sterol binding sites in Smo, but which site is endogenously regulated by Ptc remains to be determined. The potential sites of regulation include the extracellular cysteine-rich domain (CRD) of Smo, as well as a site deep within the transmembrane domain (TMD).[28][29][30]
Due to the abundance of cholesterol in the plasma membrane (up to 50 mole %), it has also been proposed that Ptc regulates the activity of Smo by controlling cholesterol accessibility specifically within the membrane of theprimary cilia,which contains a less abundant, and therefore more readily regulated pool of accessible cholesterol.[28][31]
Typically, upon activation and release of inhibition by Ptc, Smo will relocate to the primary cilia and Ptc will diffuse out of the ciliary membrane.[32]Upon inactivation, Smo no longer becomes concentrated in the ciliary membrane, This hypothesis is supported by methods which can increase or deplete the accessible cholesterol pool, with a subsequent increase or decrease in Hh signaling. This accessible cholesterol pool has been shown to be distinct from the general plasma membrane cholesterol pool in being available for protein interaction and cell uptake. The ciliary membrane has also been shown to contain lower levels of accessible cholesterol due to sequestering of cholesterol bysphingomyelin.In addition to cholesterol’s role as a Hh pathway agonist, it has been shown that cholesterol levels within the ciliary membrane rapidly increase upon treatment with Shh only in the presence of Ptc, further suggesting Ptc regulation of accessible cholesterol as the mechanism behind Smo activation/inhibition.[27]Additionally, Molecular Dynamics simulations suggest that vismodegib inhibits Smo through a conformational change that prevents cholesterol from binding.[33]This suggests the hypothesis that Ptc functions by preventing Smo access to cholesterol, and upon Ptc inhibition by Shh, Smo gains access to cholesterol and is subsequently activated, transmitting the Hh signal.
Role in disease[edit]
SMO can function as anoncogene.Activating SMO mutations can lead to unregulated activation of the hedgehog pathway and serve as driving mutations for cancers such asmedulloblastoma,basal-cell carcinoma,pancreatic cancer,andprostate cancer.[16][34]As such, SMO is an attractive cancer drug target, along with the many hedgehog pathway agonists and antagonists that are known to directly target SMO.[16]
Cholesterolis known to be crucial in regulating the overall hedgehog pathway, and congenital mutations in cholesterol synthesis pathways can inactivate SMO specifically, leading to developmental disorders.[35]For example,oxysterol20(S)-OHC is known to activate vertebrate SMO by binding the cysteine rich domain near its extracellular amino-terminal region. In the context of cancer, 20(S)-OHC is the target of a proposed anti-cancer oxysterol binding inhibitor.[18]
Agonists[edit]
- Smoothened agonist(SAG)
- Purmorphamine
- Oxysterolsincluding20(S)-OHCand20(S)-yne
Antagonists[edit]
- Cyclopamine
- Itraconazole
- Vismodegib(Erivedge), a smoothened receptor inhibitor for the treatment ofbasal-cell carcinoma,being investigated for the treatment of other types of cancer
- SANT-1
- Sonidegib
- Patidegib[36]
- Glasdegib
- Jervine
See also[edit]
References[edit]
- ^abcGRCh38: Ensembl release 89: ENSG00000128602–Ensembl,May 2017
- ^abcGRCm38: Ensembl release 89: ENSMUSG00000001761–Ensembl,May 2017
- ^"Human PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^"Mouse PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^Ruiz-Gómez A, Molnar C, Holguín H, Mayor F, de Celis JF (April 2007)."The cell biology of Smo signalling and its relationships with GPCRs".Biochimica et Biophysica Acta (BBA) - Biomembranes.1768(4): 901–12.doi:10.1016/j.bbamem.2006.09.020.hdl:10261/4737.PMID17094938.
- ^Wang C, Wu H, Evron T, Vardy E, Han GW, Huang XP, Hufeisen SJ, Mangano TJ, Urban DJ, Katritch V, Cherezov V, Caron MG,Roth BL,Stevens RC (July 2014)."Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs".Nature Communications.5:4355.Bibcode:2014NatCo...5.4355W.doi:10.1038/ncomms5355.PMC4198951.PMID25008467.
- ^Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA (August 2000). "Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine".Nature.406(6799): 1005–9.Bibcode:2000Natur.406.1005T.doi:10.1038/35023008.PMID10984056.S2CID4313790.
- ^"Vismodegib, First Hedgehog Inhibitor, Approved for BCC Patients".OncLive.Retrieved2017-05-26.
- ^Kong JH, Siebold C, Rohatgi R (May 2019)."Biochemical mechanisms of vertebrate hedgehog signaling".Development.146(10): dev166892.doi:10.1242/dev.166892.PMC6550017.PMID31092502.
- ^Jeng KS, Chang CF, Lin SS (January 2020)."Sonic Hedgehog Signaling in Organogenesis, Tumors, and Tumor Microenvironments".Int J Mol Sci.21(3): 758.doi:10.3390/ijms21030758.PMC7037908.PMID31979397.
- ^Liu X, Ding C, Tan W, Zhang A (February 2020). "Medulloblastoma: Molecular understanding, treatment evolution, and new developments".Pharmacol. Ther.210:107516.doi:10.1016/j.pharmthera.2020.107516.PMID32105673.S2CID211557426.
- ^Laliscia C, Baldaccini D, Antonuzzo A, Paiar F (2019)."Vismodegibfor the treatment of radiation-induced basal cell carcinoma - a case report and brief literature study".Contemp Oncol (Pozn).23(4): 251–253.doi:10.5114/wo.2019.91540.PMC6978755.PMID31992959.
- ^Liao S, Floyd C, Verratti N, Leung L, Wu C (March 2020)."Analysis of vismodegib resistance in D473G and W535L mutants of SMO receptor and design of novel drug derivatives using molecular dynamics simulations".Life Sci.244:117302.doi:10.1016/j.lfs.2020.117302.PMID31953165.
- ^Rohatgi R, Milenkovic L, Scott MP (July 2007)."Patched1 regulates hedgehog signaling at the primary cilium".Science.317(5836): 372–6.Bibcode:2007Sci...317..372R.doi:10.1126/science.1139740.PMID17641202.
- ^Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF (October 2005). "Vertebrate Smoothened functions at the primary cilium".Nature.437(7061): 1018–21.Bibcode:2005Natur.437.1018C.doi:10.1038/nature04117.PMID16136078.S2CID4431804.
- ^abcWang Y, Zhou Z, Walsh CT, McMahon AP (February 2009)."Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation".Proceedings of the National Academy of Sciences of the United States of America.106(8): 2623–8.Bibcode:2009PNAS..106.2623W.doi:10.1073/pnas.0812110106.PMC2650314.PMID19196978.
- ^Milenkovic L, Scott MP, Rohatgi R (November 2009)."Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium".The Journal of Cell Biology.187(3): 365–74.doi:10.1083/jcb.200907126.PMC2779247.PMID19948480.
- ^abcdArensdorf AM, Marada S, Ogden SK (January 2016)."Smoothened Regulation: A Tale of Two Signals".Trends in Pharmacological Sciences.37(1): 62–72.doi:10.1016/j.tips.2015.09.001.PMC4593303.PMID26432668.
- ^Rohatgi R, Milenkovic L, Corcoran RB, Scott MP (March 2009)."Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process".Proceedings of the National Academy of Sciences of the United States of America.106(9): 3196–201.Bibcode:2009PNAS..106.3196R.doi:10.1073/pnas.0813373106.PMC2642660.PMID19218434.
- ^Cohen MM (November 2003). "The hedgehog signaling network".American Journal of Medical Genetics. Part A.123A(1): 5–28.doi:10.1002/ajmg.a.20495.PMID14556242.S2CID31906029.
- ^Aikin RA, Ayers KL, Thérond PP (April 2008)."The role of kinases in the Hedgehog signalling pathway".EMBO Reports.9(4): 330–6.doi:10.1038/embor.2008.38.PMC2288774.PMID18379584.
- ^Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA (November 2003)."Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2".Molecular Cell.12(5): 1261–74.doi:10.1016/S1097-2765(03)00426-X.PMID14636583.
- ^Jia J, Tong C, Jiang J (November 2003)."Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail".Genes & Development.17(21): 2709–20.doi:10.1101/gad.1136603.PMC280620.PMID14597665.
- ^Hu A, Song BL (December 2019)."The interplay of Patched, Smoothened and cholesterol in Hedgehog signaling".Curr. Opin. Cell Biol.61:31–38.doi:10.1016/j.ceb.2019.06.008.PMID31369952.
- ^Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, Latorraca NR, Faust B, Dror RO, Beachy PA, Myers BR, Manglik A (July 2019)."Smoothened stimulation by membrane sterols drives Hedgehog pathway activity".Nature.571(7764): 284–288.doi:10.1038/s41586-019-1355-4.PMC6709672.PMID31263273.
- ^Kowatsch C, Woolley RE, Kinnebrew M, Rohatgi R, Siebold C (August 2019)."Structures of vertebrate Patched and Smoothened reveal intimate links between cholesterol and Hedgehog signalling".Curr. Opin. Struct. Biol.57:204–214.doi:10.1016/j.sbi.2019.05.015.PMC6744280.PMID31247512.
- ^abKinnebrew M, Iverson EJ, Patel BB, Pusapati GV, Kong JH, Johnson KA, Luchetti G, Eckert KM, McDonald JG, Covey DF, Siebold C, Radhakrishnan A, Rohatgi R (October 2019)."Cholesterol accessibility at the ciliary membrane controls hedgehog signaling".eLife.8.doi:10.7554/eLife.50051.PMC6850779.PMID31657721.
- ^abLuchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, Rohatgi R (October 2016)."Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling".eLife.5.doi:10.7554/eLife.20304.PMC5123864.PMID27705744.
- ^*Hedger G, Koldsø H, Chavent M, Siebold C, Rohatgi R, Sansom MSP (March 2019)."Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened".Structure.27(3): 549–559.e2.doi:10.1016/j.str.2018.11.003.PMC6408332.PMID30595453.
- ^Byrne EF, Luchetti G, Rohatgi R, Siebold C (April 2018)."Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates".Curr. Opin. Cell Biol.51:81–88.doi:10.1016/j.ceb.2017.10.004.PMC5949240.PMID29268141.
- ^Gigante ED, Caspary T (February 2020)."Signaling in the primary cilium through the lens of the Hedgehog pathway".Wiley Interdiscip Rev Dev Biol.9(6): e377.doi:10.1002/wdev.377.PMC7444278.PMID32084300.
- ^*Weiss LE, Milenkovic L, Yoon J, Stearns T, Moerner WE (March 2019)."Motional dynamics of single Patched1 molecules in cilia are controlled by Hedgehog and cholesterol".Proc. Natl. Acad. Sci. U.S.A.116(12): 5550–5557.Bibcode:2019PNAS..116.5550W.doi:10.1073/pnas.1816747116.PMC6431229.PMID30819883.
- ^An X, Bai Q, Bai F, Shi D, Liu H, Yao X (2019)."Deciphering the Allosteric Effect of Antagonist Vismodegib on Smoothened Receptor Deactivation Using Metadynamics Simulation".Front Chem.7:406.Bibcode:2019FrCh....7..406Y.doi:10.3389/fchem.2019.00406.PMC6558189.PMID31214579.
- ^Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, de Sauvage FJ (January 1998). "Activating Smoothened mutations in sporadic basal-cell carcinoma".Nature.391(6662): 90–2.Bibcode:1998Natur.391...90X.doi:10.1038/34201.PMID9422511.S2CID205003240.
- ^Blassberg R, Macrae JI, Briscoe J, Jacob J (February 2016)."Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome".Human Molecular Genetics.25(4): 693–705.doi:10.1093/hmg/ddv507.PMC4743690.PMID26685159.
- ^Patidegib
Further reading[edit]
- Chen Y, Struhl G (November 1996)."Dual roles for patched in sequestering and transducing Hedgehog".Cell.87(3): 553–63.doi:10.1016/S0092-8674(00)81374-4.PMID8898207.S2CID15208834.
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A (November 1996). "The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog".Nature.384(6605): 129–34.Bibcode:1996Natur.384..129S.doi:10.1038/384129a0.PMID8906787.S2CID4342540.
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, de Sauvage FJ (January 1998). "Activating Smoothened mutations in sporadic basal-cell carcinoma".Nature.391(6662): 90–2.Bibcode:1998Natur.391...90X.doi:10.1038/34201.PMID9422511.S2CID205003240.
- Reifenberger J, Wolter M, Weber RG,Megahed M,Ruzicka T, Lichter P, Reifenberger G (May 1998). "Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system".Cancer Research.58(9): 1798–803.PMID9581815.
- Sublett JE, Entrekin RE, Look AT, Reardon DA (May 1998). "Chromosomal localization of the human smoothened gene (SMOH) to 7q32. 3 by fluorescence in situ hybridization and radiation hybrid mapping".Genomics.50(1): 112–4.doi:10.1006/geno.1998.5227.PMID9628830.
- McGarvey TW, Maruta Y, Tomaszewski JE, Linnenbach AJ, Malkowicz SB (September 1998)."PTCH gene mutations in invasive transitional cell carcinoma of the bladder".Oncogene.17(9): 1167–72.doi:10.1038/sj.onc.1202045.PMID9764827.
- Chidambaram A, Gerrard B, Hanson M, Dean M (November 1998)."Chromosomal localization of the human and murine orthologues of the Drosophila smoothened gene".Genomics.53(3): 416–7.doi:10.1006/geno.1998.5531.PMID9799615.
- Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, de Sauvage FJ (November 1998)."Characterization of two patched receptors for the vertebrate hedgehog protein family".Proceedings of the National Academy of Sciences of the United States of America.95(23): 13630–4.Bibcode:1998PNAS...9513630C.doi:10.1073/pnas.95.23.13630.PMC24870.PMID9811851.
- Detmer K, Walker AN, Jenkins TM, Steele TA, Dannawi H (August 2000). "Erythroid differentiation in vitro is blocked by cyclopamine, an inhibitor of hedgehog signaling".Blood Cells, Molecules & Diseases.26(4): 360–72.doi:10.1006/bcmd.2000.0318.PMID11042037.
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP (December 2001). "Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation".Development.128(24): 5099–108.doi:10.1242/dev.128.24.5099.PMID11748145.
- Incardona JP, Gruenberg J, Roelink H (June 2002)."Sonic hedgehog induces the segregation of patched and smoothened in endosomes".Current Biology.12(12): 983–95.doi:10.1016/S0960-9822(02)00895-3.PMID12123571.S2CID3162414.
- Lowrey JA, Stewart GA, Lindey S, Hoyne GF, Dallman MJ, Howie SE, Lamb JR (August 2002)."Sonic hedgehog promotes cell cycle progression in activated peripheral CD4(+) T lymphocytes".Journal of Immunology.169(4): 1869–75.doi:10.4049/jimmunol.169.4.1869.PMID12165511.
- Taipale J, Cooper MK, Maiti T, Beachy PA (August 2002). "Patched acts catalytically to suppress the activity of Smoothened".Nature.418(6900): 892–7.Bibcode:2002Natur.418..892T.doi:10.1038/nature00989.PMID12192414.S2CID4362029.
- Katayam M, Yoshida K, Ishimori H, Katayama M, Kawase T, Motoyama J, Kamiguchi H (September 2002). "Patched and smoothened mRNA expression in human astrocytic tumors inversely correlates with histological malignancy".Journal of Neuro-Oncology.59(2): 107–15.doi:10.1023/A:1019660421216.PMID12241103.S2CID21237084.
- Couvé-Privat S, Bouadjar B, Avril MF, Sarasin A, Daya-Grosjean L (December 2002). "Significantly high levels of ultraviolet-specific mutations in the smoothened gene in basal cell carcinomas from DNA repair-deficient xeroderma pigmentosum patients".Cancer Research.62(24): 7186–9.PMID12499255.
- Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A, de Sauvage F, Dlugosz AA (June 2003)."The magnitude of hedgehog signaling activity defines skin tumor phenotype".The EMBO Journal.22(11): 2741–51.doi:10.1093/emboj/cdg271.PMC156767.PMID12773389.
- Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ (December 2004). "Activity-dependent internalization of smoothened mediated by beta-arrestin 2 and GRK2".Science.306(5705): 2257–60.Bibcode:2004Sci...306.2257C.doi:10.1126/science.1104135.PMID15618519.S2CID12823611.
- Byrne EF, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MS, Newstead S, Rohatgi R (July 2016)."Structural basis of Smoothened regulation by its extracellular domains".Nature.535(7613): 517–22.Bibcode:2016Natur.535..517B.doi:10.1038/nature18934.PMC4970916.PMID27437577.
External links[edit]
- "Frizzled Receptors: SMO".IUPHAR Database of Receptors and Ion Channels.International Union of Basic and Clinical Pharmacology. Archived fromthe originalon 2015-02-25.Retrieved2007-10-25.
- SMO+protein,+humanat the U.S. National Library of MedicineMedical Subject Headings(MeSH)





