Fusion protein

Fusion proteinsorchimeric(kī-ˈmir-ik)proteins(literally, made of parts from different sources) are proteins created through the joining of two or moregenesthat originally coded for separate proteins. Translation of thisfusion generesults in a single or multiplepolypeptideswith functional properties derived from each of the original proteins.Recombinant fusion proteinsare created artificially byrecombinant DNA technologyfor use in biological research ortherapeutics.Chimericorchimerausually designate hybrid proteins made ofpolypeptideshaving different functions or physico-chemical patterns.Chimeric mutant proteinsoccur naturally when a complexmutation,such as achromosomal translocation,tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function asoncoproteins.Thebcr-abl fusion proteinis a well-known example of an oncogenic fusion protein, and is considered to be the primary oncogenic driver ofchronic myelogenous leukemia.
Functions
[edit]Some fusion proteins combine whole peptides and therefore contain allfunctional domainsof the original proteins. However, other fusion proteins, especially those that occur naturally, combine only portions of coding sequences and therefore do not maintain the original functions of the parental genes that formed them.
Many whole gene fusions are fully functional and can still act to replace the original peptides. Some, however, experience interactions between the two proteins that can modify their functions. Beyond these effects, some gene fusions may causeregulatory changesthat alter when and where these genes act. Forpartial gene fusions,the shuffling of different active sites and binding domains have the potential to result in new proteins with novel functions.
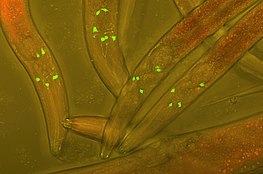
Fluorescent protein tags
[edit]The fusion offluorescent tagsto proteins in a host cell is a widely popular technique used in experimental cell and biology research in order to track protein interactions in real time. The first fluorescent tag,green fluorescent protein(GFP), was isolated fromAequorea victoriaand is still used frequently in modern research. More recent derivations include photoconvertible fluorescent proteins (PCFPs), which were first isolated fromAnthozoa.The most commonly used PCFP is theKaedefluorescent tag, but the development of Kikume green-red (KikGR) in 2005 offers a brighter signal and more efficient photoconversion. The advantage of using PCFP fluorescent tags is the ability to track the interaction of overlapping biochemical pathways in real time. The tag will change color from green to red once the protein reaches a point of interest in the pathway, and the alternate colored protein can be monitored through the duration of pathway. This technique is especially useful when studyingG-protein coupled receptor(GPCR) recycling pathways. The fates of recycled G-protein receptors may either be sent to theplasma membraneto be recycled, marked by a green fluorescent tag, or may be sent to alysosomefor degradation, marked by a red fluorescent tag.[1]
Chimeric protein drugs
[edit]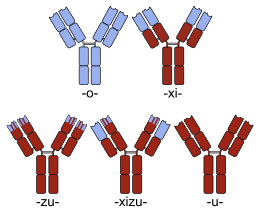
The purpose of creating fusion proteins indrug developmentis to impart properties from each of the "parent" proteins to the resulting chimeric protein. Several chimeric proteindrugsare currently available for medical use.
Many chimeric protein drugs aremonoclonal antibodieswhose specificity for atarget moleculewas developed using mice and hence were initially "mouse" antibodies. As non-human proteins, mouse antibodies tend to evoke animmune reactionif administered to humans. The chimerization process involvesengineeringthe replacement of segments of the antibody molecule that distinguish it from a human antibody. For example, humanconstant domainscan be introduced, thereby eliminating most of the potentiallyimmunogenicportions of the drug without altering its specificity for the intended therapeutic target.Antibody nomenclatureindicates this type of modification by inserting-xi-into thenon-proprietary name(e.g.,abci-xi-mab). If parts of the variable domains are also replaced by human portions,humanizedantibodies are obtained. Although not conceptually distinct from chimeras, this type is indicated using-zu-such as indacli-zu-mab.See thelist of monoclonal antibodiesfor more examples.
In addition to chimeric and humanized antibodies, there are other pharmaceutical purposes for the creation of chimeric constructs.Etanercept,for example, is aTNFαblocker created through the combination of atumor necrosis factor receptor(TNFR) with theimmunoglobulin G1Fc segment.TNFR provides specificity for the drug target and the antibody Fc segment is believed to add stability and deliverability of the drug.[2]Additional chimeric proteins used for therapeutic applications include:
- Aflibercept:A human recombinant protein that aids in the treatment of oxaliplatin-resistantmetastatic colorectal cancer,neo-vascularmacular degeneration,andmacular edema.[2]
- Rilonacept:Reduces inflammation by preventing activation ofIL-1 receptorsto treatcryopyrin-associated periodic syndromes(CAPS).[2]
- Alefacept:RegulatedT-cellresponses by selectively targeting effector memory T-cells to treatpsoriasis vulgaris.[2]
- Romiplostim:A peptibody that treatsimmune thrombocytopenia.[2]
- Abatacept/Belatacept:Interferes with T-cell co-stimulation to treat autoimmune disorders likerheumatoid arthritis,psoriatic arthritis,andpsoriasis.[2]
- Denileukin-diftitox:Treatscutaneous lymphoma.[2]
Recombinant technology
[edit]
Arecombinant fusion proteinis aproteincreated throughgenetic engineeringof a fusion gene. This typically involves removing the stopcodonfrom acDNAsequence coding for the first protein, then appending the cDNA sequence of the second proteinin framethroughligationoroverlap extension PCR.That DNA sequence will then beexpressedby acellas a single protein. The protein can be engineered to include the full sequence of both original proteins, or only a portion of either.
If the two entities are proteins, often linker (or "spacer" ) peptides are also added, which make it more likely that the proteins fold independently and behave as expected. Especially in the case where the linkers enableprotein purification,linkers in protein or peptide fusions are sometimes engineered with cleavage sites for proteases or chemical agents that enable the liberation of the two separate proteins. This technique is often used for identification and purification of proteins, by fusing aGST protein,FLAG peptide,or ahexa-his peptide(6xHis-tag), which can be isolated usingaffinity chromatographywith nickel or cobalt resins. Di- or multimeric chimeric proteins can be manufactured throughgenetic engineeringby fusion to the original proteins of peptide domains that induce artificial protein di- or multimerization (e.g.,streptavidinorleucine zippers). Fusion proteins can also be manufactured withtoxinsorantibodiesattached to them in order to study disease development. Hydrogenase promoter, PSH,was studied constructing a PSHpromoter-gfpfusion by usinggreen fluorescent protein (gfp)reporter gene.[3]
Recombinant functionality
[edit]Novel recombinant technologies have made it possible to improve fusion protein design for use in fields as diverse as biodetection, paper and food industries, and biopharmaceuticals. Recent improvements have involved the fusion of single peptides or protein fragments to regions of existing proteins, such asN and C termini,and are known to increase the following properties:[4]
- Catalytic efficiency:Fusion of certain peptides allow for greater catalytic efficiency by altering thetertiaryandquaternary structureof the target protein.[4]
- Solubility:A common challenge in fusion protein design is the issue of insolubility of newly synthesized fusion proteins in the recombinant host, leading to an over-aggregation of the target protein in the cell.Molecular chaperonesthat are able to aid in protein folding may be added, thereby better segregating hydrophobic and hydrophilic interactions in the solute to increase protein solubility.[4]
- Thermostability:Singular peptides or protein fragments are typically added to reduce flexibility of either the N or C terminus of the target protein, which reinforces thermostability and stabilizespH range.[4]
- Enzyme activity: Fusion that involves the introduction ofhydrogen bondsmay be used to expand overall enzyme activity.[4]
- Expression levels: Addition of numerous fusion fragments, such asmaltose binding protein(MBP) or small ubiquitin-like molecule (SUMO), serve to enhance enzyme expression and secretion of the target protein.[4]
- Immobilization: PHA synthase, an enzyme that allows for the immobilization of proteins of interest, is an important fusion tag in industrial research.[4]
- Crystal quality: Crystal quality can be improved by adding covalent links between proteins, aiding in structure determination techniques.[4]
Recombinant protein design
[edit]The earliest applications of recombinant protein design can be documented in the use of single peptide tags for purification of proteins inaffinity chromatography.Since then, a variety of fusion protein design techniques have been developed for applications as diverse as fluorescent protein tags to recombinant fusion protein drugs. Three commonly used design techniques include tandem fusion, domain insertion, and post-translational conjugation.[5]
Tandem fusion
[edit]The proteins of interest are simply connected end-to-end via fusion of N or C termini between the proteins. This provides a flexible bridge structure allowing enough space between fusion partners to ensure properfolding.However, the N or C termini of the peptide are often crucial components in obtaining the desired folding pattern for the recombinant protein, making simple end-to-end conjoining of domains ineffective in this case. For this reason, a protein linker is often needed to maintain the functionality of the protein domains of interest.[5]
Domain insertion
[edit]This technique involves the fusion of consecutive protein domains by encoding desired structures into a single polypeptide chain, but sometimes may require insertion of a domain within another domain. This technique is typically regarding as more difficult to carry out than tandem fusion, due to difficulty finding an appropriateligationsite in the gene of interest.[5]
Post-translational conjugation
[edit]This technique fuses protein domains following ribosomaltranslationof the proteins of interest, in contrast to genetic fusion prior to translation used in other recombinant technologies.[5]
Protein linkers
[edit]
Protein linkers aid fusion protein design by providing appropriate spacing between domains, supporting correct protein folding in the case that N or C termini interactions are crucial to folding. Commonly, protein linkers permit important domain interactions, reinforce stability, and reduce steric hindrance, making them preferred for use in fusion protein design even when N and C termini can be fused. Three major types of linkers are flexible, rigid, and in vivo cleavable.[5][6]
- Flexible linkersmay consist of many smallglycineresidues, giving them the ability curl into a dynamic, adaptable shape.[6]
- Rigid linkersmay be formed of large, cyclicprolineresidues, which can be helpful when highly specific spacing between domains must be maintained.[6]
- In vivocleavable linkersare unique in that they are designed to allow the release of one or more fused domains under certain reaction conditions, such as a specificpHgradient, or when coming in contact with anotherbiomoleculein the cell.[6]
Natural occurrence
[edit]Naturally occurring fusion genes are most commonly created when achromosomal translocationreplaces the terminalexonsof one gene with intact exons from a second gene. This creates a single gene that can betranscribed,spliced,andtranslatedto produce a functional fusion protein. Many importantcancer-promotingoncogenesare fusion genes produced in this way.
Examples include:
Antibodies are fusion proteins produced byV(D)J recombination.
There are also rare examples of naturally occurring polypeptides that appear to be a fusion of two clearly defined modules, in which each module displays its characteristic activity or function, independent of the other. Two major examples are: double PP2C chimera inPlasmodium falciparum(the malaria parasite), in which each PP2C module exhibits protein phosphatase 2C enzymatic activity,[7]and thedual-family immunophilinsthat occur in a number of unicellular organisms (such as protozoan parasites andFlavobacteria) and contain full-lengthcyclophilinandFKBPchaperone modules.[8][9]The evolutionary origin of such chimera remains unclear.
See also
[edit]References
[edit]- ^Schmidt A, Wiesner B, Schülein R, Teichmann A (2014). "Use of Kaede and Kikume Green–Red Fusions for Live Cell Imaging of G Protein-Coupled Receptors".Exocytosis and Endocytosis.Methods in Molecular Biology. Vol. 1174. New York, NY: Humana Press. pp. 139–156.doi:10.1007/978-1-4939-0944-5_9.ISBN9781493909438.PMID24947379.
- ^abcdefgBaldo BA (May 2015). "Chimeric fusion proteins used for therapy: indications, mechanisms, and safety".Drug Safety.38(5): 455–79.doi:10.1007/s40264-015-0285-9.PMID25832756.S2CID23852865.
- ^Jugder BE, Welch J, Braidy N, Marquis CP (2016-07-26)."Construction and use of a Cupriavidus necator H16 soluble hydrogenase promoter (PSH) fusion to gfp (green fluorescent protein)".PeerJ.4:e2269.doi:10.7717/peerj.2269.PMC4974937.PMID27547572.
- ^abcdefghYang H, Liu L, Xu F (October 2016). "The promises and challenges of fusion constructs in protein biochemistry and enzymology".Applied Microbiology and Biotechnology.100(19): 8273–81.doi:10.1007/s00253-016-7795-y.PMID27541749.S2CID14316893.
- ^abcdeYu K, Liu C, Kim BG, Lee DY (2015-01-01). "Synthetic fusion protein design and applications".Biotechnology Advances.33(1): 155–164.doi:10.1016/j.biotechadv.2014.11.005.PMID25450191.
- ^abcdChen X, Zaro JL, Shen WC (October 2013)."Fusion protein linkers: property, design and functionality".Advanced Drug Delivery Reviews.65(10): 1357–69.doi:10.1016/j.addr.2012.09.039.PMC3726540.PMID23026637.
- ^Mamoun CB, Sullivan DJ, Banerjee R, Goldberg DE (May 1998)."Identification and characterization of an unusual double serine/threonine protein phosphatase 2C in the malaria parasite Plasmodium falciparum".The Journal of Biological Chemistry.273(18): 11241–7.doi:10.1074/jbc.273.18.11241.PMID9556615.
- ^Adams B, Musiyenko A, Kumar R, Barik S (July 2005)."A novel class of dual-family immunophilins".The Journal of Biological Chemistry.280(26): 24308–14.doi:10.1074/jbc.M500990200.PMC2270415.PMID15845546.
- ^Barik S (November 2017)."On the role, ecology, phylogeny, and structure of dual-family immunophilins".Cell Stress & Chaperones.22(6): 833–845.doi:10.1007/s12192-017-0813-x.PMC5655371.PMID28567569.
External links
[edit]- Mutant+Chimeric+Proteinsat the U.S. National Library of MedicineMedical Subject Headings(MeSH)
- ChiPPIArchived2021-11-10 at theWayback Machine:The Server Protein–Protein Interaction of Chimeric Proteins.
