Apusomonadidae
| Apusomonadidae | |
|---|---|
| |
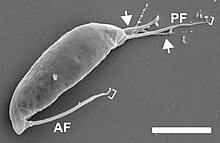
| Podomonas kaiyoaeSEMimage. AF: anteriorflagellum.PF: posterior flagellum. T: tusk. Brackets:acronemes.Arrows and arrowheads:pseudopodium.Scale bar = 5 μm. | |
| Scientific classification | |
| Domain: | Eukaryota |
| Clade: | Amorphea |
| Clade: | Obazoa |
| Class: | Thecomonadea Cavalier-Smith, 1993 emend. 2013[2] |
| Order: | Apusomonadida Karpov & Mylnikov, 1989[1] |
| Family: | Apusomonadidae Karpov & Mylnikov, 1989[1] |
| Genera | |
|
See text | |
| Diversity | |
| 28 species | |
Theapusomonads(familyApusomonadidae) are a group ofprotozoanzooflagellatesthat glide on surfaces, and mostly consumeprokaryotes.They are of particular evolutionary interest because they appear to be the sister group to theOpisthokonts,the clade that includes bothanimalsandfungi.Together with theBreviatea,these form theObazoaclade.[3][4][5]
Characteristics
[edit]Apusomonads are smallglidingheterotrophicbiflagellates(i.e. with twoflagella) that possess aproboscis,formed partly or entirely by the anterior flagellum surrounded by a membranous sleeve. There is apellicleunder the dorsal cell membrane that extends into the proboscis sleeve and into a skirt that covers the sides of the cell. Apusomonads present two different cell plans:[6]
- Derived cell plan, represented byApusomonas,with a roundcell bodyand amastigophore,a projection of the cell containing bothbasal bodiesat its end.[6]
- "Amastigomonas-like "cell plan, with an oval or oblong cell that generally formspseudopodiafrom the ventral surface, with no mastigophore, and the proboscis comprising solely the flagellum and the sleeve. These characteristics are considered 'primitive' or 'ancestral' in comparison withApusomonas.Organisms with this body plan, although historically assigned to the same genusAmastigomonas,are aparaphyleticgroup from whichApusomonashas evolved.[6][7]
Evolution
[edit]External relationships
[edit]The apusomonads are the sister group toOpisthokonta,the lineage that includesanimals,fungiand an array of relatedprotists.Because of this, apusomonads occupy an importantphylogeneticposition to understandeukaryotic evolution.They retain ancestral characteristics, such as thebiflagellatebody plan,which in opisthokonts evolves into a uniflagellate plan.[7]
Apusomonads are vital to understandingmulticellularity.Genes involved in multicellularity have been found in the apusomonadThecamonas,[8]such as adhesion proteins, calcium-signaling genes and types of sodium channels characteristic of animals.[6]Thegenomeof the strain "Amastigomonassp. "presents theintegrin-mediatedadhesionmachinery, the primarycell-matrix adhesionmechanism seen inMetazoa(animals).[9]
Internal relationships
[edit]Apusomonads are a poorly and narrowly studied group.[6]Currently, the diversity of described apusomonads consists of the roundApusomonasand a wide array of "Amastigomonas-type "organisms that have been reclassified into the generaThecamonas,Manchomonas,Podomonas,Multimonas,Chelonemonasand, most recently,Catacumbia,Cavaliersmithia,Karpovia,MylnikoviaandSingekia.The relationships between these genera are depicted by the cladogram below.[7]
| Apusomonadida | "Amastigomonas-like "organisms | |
Taxonomy
[edit]History
[edit]Apusomonads were first described in 1989 as onefamilyApusomonadidaeinside the monotypicorderApusomonadida,as a group of flagellates containing the generaApusomonasandAmastigomonas.[1]Later, BritishprotozoologistThomas Cavalier-Smithclassified them within the monotypicclassThecomonadeaas part of theparaphyleticphylumApusozoa.[2]Moderncladisticapproaches toeukaryoticclassification refer to apusomonads by their order-level name alone.[7][10]
Classification
[edit]There are 10 recognized genera, as well as the "Amastigomonas-like "archetypethat includes primitive forms not yet transferred to new genera.[7]
- Amastigomonasde Saedeleer 1931
- A. caudataMylnikov 1989[Amastigomonas borokensisHamar 1979]
- A. debruyneide Saedeleer 1931
- A. marisrubriMylnikov & Mylnikov 2012
- CatacumbiaTorruella, Galindo et al. 2022[7]
- C. lutetiensisTorruella, Galindo et al. 2022
- CavaliersmithiaTorruella, Galindo et al. 2022
- C. chaoaeTorruella, Galindo et al. 2022
- MultimonasCavalier-Smith 2010
- M. koreensisHeiss, Lee, Ishida & Simpson, 2015
- M. marina(Mylnikov 1989) Cavalier-Smith 2010[Cercomonas marinaMylnikov 1989;Amastigomonas marina(Mylnikov 1989) Mylnikov 1999]
- M. mediaCavalier-Smith 2010
- MylnikoviaTorruella, Galindo et al. 2022
- M. oxoniensis(Cavalier-Smith 2010) Torruella, Galindo et al. 2022[Thecamonas oxoniensisCavalier-Smith 2010]
- PodomonasCavalier-Smith 2010
- P. capensisCavalier-Smith 2010
- P. gigantea(Mylnikov 1999)[Amastigomonas giganteaMylnikov 1999]
- P. griebenis(Mylnikov 1999)[Amastigomonas griebenisMylnikov 1999]
- P. kaiyoaeYabuki in Yabuki, Tame & Mizuno 2022[11]
- P. klosteris(Arndt & Mylnikov 1999) Cavalier-Smith 2010[Amastigomonas klosterisArndt & Mylnikov 1999]
- P. magnaCavalier-Smith 2010
- ApusomonadinaeCavalier-Smith 2010[3]
- ApusomonasAlexeieff 1924[RostromonasKarpoff & Zhukov 1980]
- A. australiensisEkelund & Patterson 1997
- A. proboscideaAlexeieff 1924[Rostromonas applanataKarpoff & Zhukov 1980]
- ManchomonasCavalier-Smith 2010
- M. bermudensis(Molina & Nerad 1991) Cavalier-Smith 2010[Amastigomonas bermudensisMolina & Nerad 1991]
- ApusomonasAlexeieff 1924[RostromonasKarpoff & Zhukov 1980]
- ThecamonadinaeLarsen & Patterson 1990[Thecamonas/Chelomonasclade]
- ChelonemonasHeiss, Lee, Ishida & Simpson, 2015
- C. dolaniTorruella, Galindo et al. 2022
- C. geobukHeiss, Lee, Ishida & Simpson, 2015
- C. masanensisHeiss, Lee, Ishida & Simpson, 2015
- KarpoviaTorruella, Galindo et al. 2022
- K. croaticaTorruella, Galindo et al. 2022
- SingekiaTorruella, Galindo et al. 2022
- S. franciliensisTorruella, Galindo et al. 2022
- S. montserratensisTorruella, Galindo et al. 2022
- ThecamonasLarsen & Patterson 1990
- T. filosaLarsen & Patterson 1990[Amastigomonas filosa(Larsen & Patterson 1990) Molina & Nerad 1991]
- T. muscula(Mylnikov 1999) Cavalier-Smith 2010[Amastigomonas musculaMylnikov 1999]
- T. mutabilis(Griessmann 1913) Larsen & Patterson 1990[Rhynchomonas mutabilisGriessmann 1913;Amastigomonas mutabilis(Griessmann 1913) Patterson & Zölffel 1993]
- T. trahensLarsen & Patterson 1990[Amastigomonas trahens(Larsen & Patterson 1990) Molina & Nerad 1991]
- ChelonemonasHeiss, Lee, Ishida & Simpson, 2015
References
[edit]- ^abcKarpov SA, Mylnikov AP (1989)."БИОЛОГИЯ И УЛЬТРАСТРУКТУРА БЕСЦВЕТНЫХ ЖГУТИКОНОСЦЕВ APUSOMONADIDA ORD.N"[Biology and ultrastructure of colourless flagellates Apusomonadida ord. n.](PDF).Zoologischkeiĭ Zhurnal(in Russian).LXVIII(8): 5–17.
- ^abCavalier-Smith, Thomas (May 2013). "Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa".European Journal of Protistology.49(2): 115–178 Document online.doi:10.1016/j.ejop.2012.06.001.ISSN0932-4739.PMID23085100.
- ^abCavalier-Smith, Thomas; Chao, Ema E. (October 2010). "Phylogeny and evolution of Apusomonadida (Protozoa: Apusozoa): new genera and species".Protist.161(4): 549–576.doi:10.1016/j.protis.2010.04.002.PMID20537943.
- ^Brown, Matthew W.; Sharpe, Susan C.; Silberman, Jeffrey D.; Heiss, Aaron A.; Lang, B. Franz; Simpson, Alastair G. B.; Roger, Andrew J. (2013-10-22)."Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads".Proceedings of the Royal Society of London B: Biological Sciences.280(1769): 20131755.doi:10.1098/rspb.2013.1755.ISSN0962-8452.PMC3768317.PMID23986111.
- ^Eme, Laura; Sharpe, Susan C.; Brown, Matthew W.; Roger, Andrew J. (2014-08-01)."On the Age of Eukaryotes: Evaluating Evidence from Fossils and Molecular Clocks".Cold Spring Harbor Perspectives in Biology.6(8): a016139.doi:10.1101/cshperspect.a016139.ISSN1943-0264.PMC4107988.PMID25085908.
- ^abcdeHeiss AA, Lee WJ, Ishida KI, Simpson AG (2015). "Cultivation and Characterisation of New Species of Apusomonads (the Sister Group to Opisthokonts), Including Close Relatives ofThecamonas(Chelonemonasn. gen.) ".Journal of Eukaryotic Microbiology.62(5): 637–649.doi:10.1111/jeu.12220.PMID25912654.
- ^abcdefTorruella G, Galindo LJ, Moreira D, Ciobanu M, Heiss AA, Yubuki N, et al. (November 2022). "Expanding the molecular and morphological diversity of Apusomonadida, a deep-branching group of gliding bacterivorous protists".Journal of Eukaryotic Microbiology.70(2): e12956.doi:10.1111/jeu.12956.hdl:2117/404026.
- ^Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I (2010)."Ancient origin of the integrin-mediated adhesion and signaling machinery".Proceedings of the National Academy of Sciences of the United States of America.107(22): 10142–10147.doi:10.1073/pnas.1002257107.PMC2890464.
- ^Sebé-Pedrós A, Ruiz-Trillo I (2010)."Integrin-mediated adhesion complex".Communicative & Integrative Biology.3(5): 475–477.doi:10.4161/cib.3.5.12603.PMC2974085.
- ^Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019)."Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes".Journal of Eukaryotic Microbiology.66(1): 4–119.doi:10.1111/jeu.12691.PMC6492006.PMID30257078.
- ^Yabuki A, Tame A, Mizuno K (2022)."Podomonas kaiyoae n. sp., a novel apusomonad growing axenically".Journal of Eukaryotic Microbiology.00:e12946.doi:10.1111/jeu.12946.
