Biomarkers of aging
Biomarkers of agingarebiomarkersthat could predict functional capacity at some later age better than chronological age.[1]Stated another way, biomarkers ofagingwould give the true "biological age", which may be different from the chronological age.
Validated biomarkers of aging would allow for testing interventions toextend lifespan,because changes in the biomarkers would be observable throughout the lifespan of the organism.[1]Althoughmaximum lifespanwould be a means of validating biomarkers of aging, it would not be a practical means for long-lived species such as humans becauselongitudinal studieswould take far too much time.[2]Ideally, biomarkers of aging should assay the biological process ofagingand not a predisposition to disease, should cause a minimal amount of trauma to assay in the organism, and should be reproducibly measurable during a short interval compared to the lifespan of the organism.[1]An assemblage of biomarker data for an organism could be termed its "ageotype".[3]
Althoughgraying of hairincreases with age,[4]hair graying cannot be called a biomarker of ageing. Similarly,skin wrinklesand other common changes seen with aging are not better indicators of future functionality than chronological age.Biogerontologistshave continued efforts to find and validate biomarkers of aging, but success thus far has been limited. Levels ofCD4and CD8memory T cellsandnaive T cellshave been used to give good predictions of the expected lifespan of middle-aged mice.[5]
Advances inbig dataanalysis allowed for the new types of "aging clocks" to be developed. Theepigenetic clockis a promising biomarker of aging and can accurately predict human chronological age.[6]Basic blood biochemistry and cell counts can also be used to accurately predict the chronological age.[7]Further studies of thehematological clockon the large datasets from South Korean, Canadian, and Eastern European populations demonstrated that biomarkers of aging may be population-specific and predictive of mortality.[8] It is also possible to predict the human chronological age using thetranscriptomic clock.[9]
Epigenetic marks
[edit]Loss of histones
[edit]A newepigeneticmark found in studies of aging cells is the loss ofhistones.Most of the evidence shows that loss of histones is linked to cell division. In aging and dividingyeastMNase-seq (Micrococcal Nuclease sequencing) showed a loss of nucleosomes of ~50%. Proper histone dosage is important in yeast as shown from the extended lifespans seen in strains that are overexpressing histones.[10]A consequence of histone loss in yeast is the amplification oftranscription.In younger cells, genes that are most induced with age have specific chromatin structures, such as fuzzy nuclear positioning, lack of anucleosomedepleted region (NDR) at thepromoter,weak chromatin phasing, a higher frequency ofTATA elements,and higher occupancy of repressive chromatin factors. In older cells, however, the same genes nucleosome loss at the promoter is more prevalent which leads to higher transcription of these genes.[10]
This phenomenon is not only seen in yeast, but has also been seen in aging worms, during aging of human diploid primaryfibroblasts,and insenescenthuman cells. In human primary fibroblasts, reduced synthesis of new histones was seen to be a consequence of shortenedtelomeresthat activate the DNA damage response. Loss of core histones may be a general epigenetic mark of aging across many organisms.[11]
Histone variants
[edit]In addition to the core histones, H2A, H2B, H3, and H4, there are other versions of the histone proteins that can be significantly different in their sequence and are important for regulating chromatin dynamics. Histone H3.3 is a variant of histone H3 that is incorporated into the genome independent of replication. It is the major form of histone H3 seen in the chromatin of senescent human cells, and it appears that excess H3.3 can drivesenescence.[11]
There are multiple variants of histone 2, the one most notably implicated in aging is macroH2A. The function of macroH2A has generally been assumed to be transcriptional silencing; most recently, it has been suggested that macroH2A is important in repressing transcription at Senescence-Associated Heterochromatin Foci (SAHF).[11]Chromatin that contains macroH2A is impervious to ATP-dependent remodeling proteins and to the binding oftranscription factors.[12]
Histone modifications
[edit]Increasedacetylation of histonescontributes to chromatin taking a moreeuchromaticstate as an organism ages, similar to the increasedtranscriptionseen due to the loss of histones.[13]There is also a reduction in the levels of H3K56ac during aging and an increase in the levels ofH4K16ac.[10]Increased H4K16ac in old yeast cells is associated with the decline in levels of theHDACSir2, which can increase the life span when overexpressed.[10]
Methylation of histoneshas been tied to life span regulation in many organisms, specifically H3K4me3, an activating mark, and H4K27me3, a repressing mark. InC. elegans,the loss of any of the three Trithorax proteins that catalyze the trimethylation of H3K4 such as, WDR-5 and the methyltransferases SET-2 and ASH-2, lowers the levels of H3K4me3 and increases lifespan. Loss of the enzyme that demethylates H3K4me3, RB-2, increases H3K4me3 levels inC. elegansand decreases their life spans.[13]In therhesus macaquebrainprefrontal cortex,H3K4me2increases atpromotersandenhancersduring postnatal development andaging.[14]These increases reflect progressively more active andtranscriptionallyaccessible (or open)chromatinstructures that are often associated withstressresponses such as theDNA damageresponse. These changes may form anepigeneticmemory of stresses and damages experienced by the organism as it develops and ages.[14]
UTX-1, aH3K27me3demethylase, plays a critical role in the aging ofC.elegans:increasedutx-1expression correlates with a decrease in H3K27me3 and a decrease in lifespan.Utx-1knockdowns showed an increase in lifespan[13]Changes in H3K27me3 levels also have affects on aging cells inDrosophilaand humans.
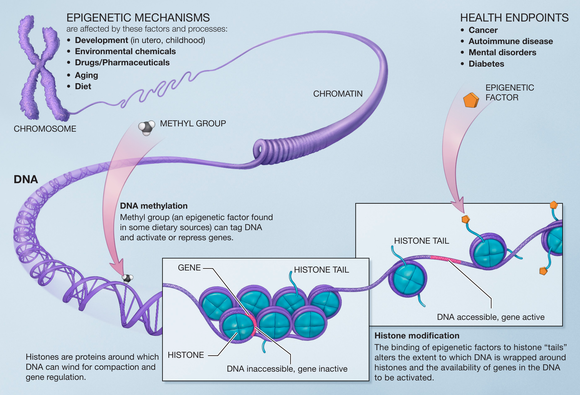
DNA methylation
[edit]Methylation of DNAis a common modification inmammaliancells. Thecytosinebase is methylated and becomes5-methylcytosine,most often when in theCpGcontext. Hypermethylation ofCpG islandsis associated with transcriptional repression and hypomethylation of these sites is associated with transcriptional activation. Many studies have shown that there is a loss of DNA methylation during ageing in many species such as, rats, mice, cows, hamsters, and humans. It has also been shown thatDNMT1andDNMT3adecrease with aging andDNMT3bincreases.[15]
Hypomethylation of DNA can lower genomic stability, induce the reactivation oftransposable elements,and cause the loss ofimprinting,all of which can contribute tocancerprogression andpathogenesis.[15]
Immune biomarkers
[edit]Recent data suggests that an increased frequency of senescent CD8+ T cells in the peripheral blood is associated with the development of hyperglycemia from a pre-diabetic state suggestive of senescence playing a role in metabolic aging. Senescent Cd8+ T cells could be utilized as a biomarker to signal the transition from pre-diabetes to overt hyperglycemia.[16]
Recently, Hashimoto and coworkers profiled thousands of circulating immune cells from supercentenarians at single-cell resolution. They identified a unique increase in cytotoxic CD4 T cells in these supercentenarians. Generally, CD4 T-cells have helper, but not cytotoxic, functions under physiological conditions however these supercentenarians, subjected to single cell profiling of their T-cell receptors, revealed accumulations of cytotoxic CD4 T-cells through clonal expansion. The conversion of helper CD4 T-cells to a cytotoxic variety might be an adaptation to the late stage of aging aiding in the fighting infections and potentially enhancing tumor surveillance.[17]
Applications of aging biomarkers
[edit]The main mechanisms identified as potential biomarkers of aging are DNA methylation, loss of histones, and histone modification. The uses for biomarkers of aging are ubiquitous and identifying a physical parameter of biological aging would allow humans to determine our true age, mortality, and morbidity.[10]The change in the physical biomarker should be proportional to the change in the age of the species. Thus after establishing a biomarker of aging, humans would be able to dive into research on extending life spans and finding timelines for the arise of potential genetic diseases.
One of the applications of this finding would allow for identification of the biological age of a person. DNA methylation uses the structure of dna at different stages of life to determine an age. DNA methylation is the methylation of the cysteine in the CG or Cpg region. The hypermethylation of this region is associated with decreased transcriptional activity and the opposite for hypomethylation. In other words, the more "tightly" held the DNA region then the more stable and "younger" the species. Looking at DNA methylation's properties in tissues, it was found to be almost zero for embryonic tissues, it can be used to determine acceleration of age and the results can be reproduced in chimpanzee tissue.[18]
More recently, biomarkers of aging has been used in multiple clinical trials to measure slowing or reversing of age-related decline or biological aging.[19]The Biomarkers of Aging Consortium (https:// agingconsortium.org) is currently examining the application of these biomarkers to identify longevity interventions and ways to validate them.[20]Moreover, open-source resources, such as the R package methylCIPHER[21]and the Python package pyaging[22]are available to the public as hubs for several biomarkers of aging.
See also
[edit]References
[edit]- ^abcBaker GT, Sprott RL (1988)."Biomarkers of aging".Experimental Gerontology.23(4–5): 223–39.doi:10.1016/0531-5565(88)90025-3.PMID3058488.S2CID31039588.
- ^Harrison, Ph.D., David E. (November 11, 2011)."V. Life span as a biomarker".Jackson Laboratory.Archived fromthe originalon April 26, 2012.Retrieved2011-12-03.
- ^Ahadi, Sara; Zhou, Wenyu; Schüssler-Fiorenza Rose, Sophia Miryam; Sailani, M. Reza; Contrepois, Kévin; Avina, Monika; Ashland, Melanie; Brunet, Anne; Snyder, Michael (2020)."Personal aging markers and ageotypes revealed by deep longitudinal profiling".Nature Medicine.26(1): 83–90.doi:10.1038/s41591-019-0719-5.PMC7301912.PMID31932806.
- ^Van Neste D, Tobin DJ (2004). "Hair cycle and hair pigmentation: dynamic interactions and changes associated with aging".Micron.35(3): 193–200.doi:10.1016/j.micron.2003.11.006.PMID15036274.
- ^Miller RA (April 2001)."Biomarkers of aging: prediction of longevity by using age-sensitive T-cell subset determinations in a middle-aged, genetically heterogeneous mouse population".The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences.56(4): B180-6.doi:10.1093/gerona/56.4.b180.PMC7537444.PMID11283189.
- ^Horvath S (2013)."DNA methylation age of human tissues and cell types".Genome Biology.14(10): R115.doi:10.1186/gb-2013-14-10-r115.PMC4015143.PMID24138928.(Erratum:doi:10.1186/s13059-015-0649-6,PMID25968125,Retraction Watch)
- ^Putin E, Mamoshina P, Aliper A, Korzinkin M, Moskalev A, Kolosov A, Ostrovskiy A, Cantor C, Vijg J, Zhavoronkov A (May 2016)."Deep biomarkers of human aging: Application of deep neural networks to biomarker development".Aging.8(5): 1021–33.doi:10.18632/aging.100968.PMC4931851.PMID27191382.
- ^Mamoshina P, Kochetov K, Putin E, Cortese F, Aliper A, Lee WS, Ahn SM, Uhn L, Skjodt N, Kovalchuk O, Scheibye-Knudsen M, Zhavoronkov A (October 2018)."Population Specific Biomarkers of Human Aging: A Big Data Study Using South Korean, Canadian, and Eastern European Patient Populations".The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences.73(11): 1482–1490.doi:10.1093/gerona/gly005.PMC6175034.PMID29340580.
- ^Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, et al. (October 2015)."The transcriptional landscape of age in human peripheral blood".Nature Communications.6:8570.Bibcode:2015NatCo...6.8570..doi:10.1038/ncomms9570.PMC4639797.PMID26490707.
- ^abcdeSen P, Shah PP, Nativio R, Berger SL (August 2016)."Epigenetic Mechanisms of Longevity and Aging".Cell.166(4): 822–839.doi:10.1016/j.cell.2016.07.050.PMC5821249.PMID27518561.
- ^abcPal S, Tyler JK (July 2016)."Epigenetics and aging".Science Advances.2(7): e1600584.Bibcode:2016SciA....2E0584P.doi:10.1126/sciadv.1600584.PMC4966880.PMID27482540.
- ^Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, et al. (January 2005)."Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA".Developmental Cell.8(1): 19–30.doi:10.1016/j.devcel.2004.10.019.PMID15621527.
- ^abcMcCauley BS, Dang W (December 2014)."Histone methylation and aging: lessons learned from model systems".Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms.1839(12): 1454–62.doi:10.1016/j.bbagrm.2014.05.008.PMC4240748.PMID24859460.
- ^abHan Y, Han D, Yan Z, Boyd-Kirkup JD, Green CD, Khaitovich P, Han JD (December 2012)."Stress-associated H3K4 methylation accumulates during postnatal development and aging of rhesus macaque brain".Aging Cell.11(6): 1055–64.doi:10.1111/acel.12007.PMID22978322.S2CID17523080.
- ^abLillycrop KA,Hoile SP, Grenfell L, Burdge GC (August 2014)."DNA methylation, ageing and the influence of early life nutrition".The Proceedings of the Nutrition Society.73(3): 413–21.doi:10.1017/S0029665114000081.PMID25027290.
- ^Lee, Yong-ho; Kim, So Ra; Han, Dai Hoon; Yu, Hee Tae; Han, Yoon Dae; Kim, Jin Hee; Kim, Soo Hyun; Lee, Chan Joo; Min, Byoung-Hoon; Kim, Dong-Hyun; Kim, Kyung Hwan (2018-11-02)."Senescent T Cells Predict the Development of Hyperglycemia in Humans".Diabetes.68(1): 156–162.doi:10.2337/db17-1218.ISSN0012-1797.PMID30389747.
- ^Hashimoto, Kosuke; Kouno, Tsukasa; Ikawa, Tomokatsu; Hayatsu, Norihito; Miyajima, Yurina; Yabukami, Haruka; Terooatea, Tommy; Sasaki, Takashi; Suzuki, Takahiro (2019-05-20)."Single-cell transcriptomics reveals expansion of cytotoxic CD4 T-cells in supercentenarians".Proceedings of the National Academy of Sciences of the United States of America.116(48): 24242–24251.Bibcode:2019PNAS..11624242H.bioRxiv10.1101/643528.doi:10.1073/pnas.1907883116.PMC6883788.PMID31719197.
- ^Horvath, Steve (2013)."DNA methylation age of human tissues and cell types".Genome Biology.14(10): R115.doi:10.1186/gb-2013-14-10-r115.ISSN1465-6906.PMC4015143.PMID24138928.(Erratum:doi:10.1186/s13059-015-0649-6,PMID25968125,Retraction Watch)
- ^Moqri, Mahdi; Herzog, Chiara; Poganik, Jesse R.; Justice, Jamie; Belsky, Daniel W.; Higgins-Chen, Albert; Moskalev, Alexey; Fuellen, Georg; Cohen, Alan A.; Bautmans, Ivan; Widschwendter, Martin; Ding, Jingzhong; Fleming, Alexander; Mannick, Joan; Han, Jing-Dong Jackie; Zhavoronkov, Alex; Barzilai, Nir; Kaeberlein, Matt; Cummings, Steven; Kennedy, Brian K.; Ferrucci, Luigi; Horvath, Steve; Verdin, Eric; Maier, Andrea B.; Snyder, Michael P.; Sebastiano, Vittorio; Gladyshev, Vadim N.; Gladyshev, V. N. (2023)."Biomarkers of aging for the identification and evaluation of longevity interventions".Cell.186(18): 3758–3775.doi:10.1016/j.cell.2023.08.003.PMC11088934.PMID37657418.
- ^Moqri, Mahdi; Herzog, Chiara; Poganik, Jesse R.; Ying, Kejun; Justice, Jamie N.; Belsky, Daniel W.; Higgins-Chen, Albert T.; Chen, Brian H.; Cohen, Alan A.; Fuellen, Georg; Hägg, Sara; Marioni, Riccardo E.; Widschwendter, Martin; Fortney, Kristen; Fedichev, Peter O. (February 2024)."Validation of biomarkers of aging".Nature Medicine.30(2): 360–372.doi:10.1038/s41591-023-02784-9.ISSN1546-170X.PMC11090477.PMID38355974.
- ^Thrush, Kyra L.; Higgins-Chen, Albert T.; Liu, Zuyun; Levine, Morgan E. (2022)."bioRxiv".doi:10.1101/2022.07.13.499978.
{{cite journal}}:Cite journal requires|journal=(help) - ^Camillo, Lucas Paulo de Lima (2024)."pyaging: a Python-based compendium of GPU-optimized aging clocks".Bioinformatics.40(btae200): btae200.doi:10.1093/bioinformatics/btae200.PMC11058068.PMID38603598.
