Deep homology

Inevolutionary developmental biology,the concept ofdeep homologyis used to describe cases wheregrowthanddifferentiationprocesses are governed by genetic mechanisms that arehomologousand deeplyconservedacross a wide range ofspecies.
History[edit]
In 1822, the French zoologistÉtienne Geoffroy Saint-Hilairedissected acrayfish,discovering that its body is organised like a vertebrate's, butinverted belly to back (dorsoventrally):[1]
I just found that all the soft organs, that is to say, the principal organs of life are found in crustaceans, and so in insects, in the same order, in the same relationships and with the same arrangement as their analogues in the high vertebrate animals... What was my surprise, and I may add, my admiration, seeing [such] a rule...[1]
Geoffroy's homology theory was denounced by the leading French zoologist of his day,Georges Cuvier,but in 1994, Geoffroy was shown to be correct.[1]In 1915,Santiago Ramon y Cajalmapped the neural connections of the optic lobes of a fly, finding that these resembled those of vertebrates.[1]In 1978,Edward B. Lewishelped to foundevolutionary developmental biology,discovering thathomeotic genesregulated embryonic development in fruit flies.[1]
In 1997, the term deep homology first appeared in a paper byNeil Shubin,Cliff Tabin, andSean B. Carroll,describing the apparent relatedness in genetic regulatory apparatuses which indicated evolutionary similarities in disparate animal features.[2]
Difference from ordinary homology[edit]
Whereas ordinaryhomologyis seen in the pattern of structures such as limb bones of mammals that are evidently related, deep homology can apply to groups of animals that have quite dissimilar anatomy: vertebrates (withendoskeletonsmade ofboneandcartilage) and arthropods (withexoskeletonsmade ofchitin) nevertheless have limbs that are constructed using similar recipes or "algorithms".[2][3][4][5]
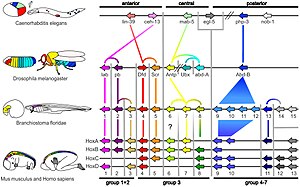
Within themetazoa,homeotic genescontrol differentiation along majorbody axes,andpax genes(especiallyPAX6) help to control the development of theeyeand othersensory organs.The deep homology applies across widely separated groups, such as in the eyes ofmammalsand the structurally quite differentcompound eyesofinsects.[3]
Similarly,hox geneshelp to form an animal's segmentation pattern. HoxA and HoxD, that regulate finger and toe formation in mice, control the development ofray finsinzebrafish;these structures had until then been considered non-homologous.[6]
There is a possible deep homology among animals that use acoustic communication, such as songbirds and humans, which may share unmutated versions of theFOXP2gene.[7]
In cancer stem cells[edit]
In modern daybiology,the depth of understanding deep homology has evolved into focusing on themolecularandgenetic mechanismsand functions rather than simplemorphology.Cancer stem cells(CSCs) are a population of cells within a tumor that have the ability to self-renew and differentiate into different cell types, similar to normalstem cells.The stem cell theory of cancer suggests that there is a subpopulation of cells, referred to as cancer stem cells, that have certain characteristics that make them unique among other types of cells within a cancer. The traits that are included in CSCs are that they multiply indefinitely, are resistant tochemotherapy,and are proposed to be responsible for relapse after therapy.[8]
Life cycle of cancer[edit]
Theunicellular life cycleof cancer andEntamoebais uniquely similar, and thus contradicts themolecular phylostratigraphic theoryfor the origin of cancer. This deep relationship between the two cell systems is supported by the "amoeba model", which provides a greater understanding of the biology of cancer from the evolutionary perspective.[9]The G + S life cycle ofEntamoebais the closest common ancestor than compared to any other life cycle of unicellular organisms. Similarly, both cell systems,amoebaandcancer,use the deep homologous G + S gene module that was evolved by a common ancestor. Some parallels that they share are too close for coincidence including:
- A reproductive asexual germ-line capable of forminggerm-line stem cells(GSCs, referred to as CSCs in cancer) and a somatic cell line without reproductive GSC function;
- Germandsoma cellsthat proliferate through asymmetric and symmetric cell cycles and can interconvert by transitioning from germ to soma (GST) and from soma to germ (SGT); both processes are referred to as MET and EMT in cancer;
- Oxygen-sensitive germlines that irreversibly lose their reproductive function due to irreparable DNA damage caused by excess oxygen;
- DNA damage repair (DDR) mechanisms to repairDNA replicationandpolyploidizationdefects and maintain genomic integrity of nascent GSCs/CSCs;
- DNA DSB repair mechanisms via MGRS and PGCC structures, with or withouthomologous cell fusion.[9]
MGRSs are also known in medical terms as “pre-existing Polypoid Giant Cancer Cells (PGCCs)” and are frequently observed in untreated cancers.[citation needed]In cancer, the reproductivegerm-linecycle starts with a precursor cell. This cell will then polyploidize within a cell envelope. This cancer germ-line undergoes a process of development that is similar to the Entamoeba germline. A significant trace of deep homology can be found in mammalian germ-line stem cells. Based on a previous hypothesis, the germ-line is the common ancestor in somatic stem cell lineages. Daughter GSCs are the only stem cells that have the capability of passing genetic information throughout generations.[9]
Algorithm[edit]
In 2010, a team led byEdward Marcottedeveloped analgorithmthat identifies deeply homologous genetic modules in unicellular organisms, plants, and animals based onphenotypes(such as traits and developmental defects). The technique aligns phenotypes across organisms based onorthology(a type of homology) of genes involved in the phenotypes.[10][11]
See also[edit]
- Body plan– Set of morphological features common to members of a phylum of animals
References[edit]
- ^abcdeHeld, Lewis I.(February 2017).Deep Homology?: Uncanny Similarities of Humans and Flies Uncovered by Evo-Devo.Cambridge University Press. pp. 2–5.ISBN978-1316601211.
- ^abShubin, Neil; Tabin, Cliff; Carroll, Sean (1997)."Fossils, genes and the evolution of animal limbs".Nature.388(6643). Springer Nature: 639–648.Bibcode:1997Natur.388..639S.doi:10.1038/41710.PMID9262397.S2CID2913898.
- ^abCarroll, Sean B.(2006).Endless Forms Most Beautiful.Weidenfeld & Nicolson. pp. 28, 66–69.ISBN0-297-85094-6.
- ^Gilbert, Scott F.(2000)."Homologous Pathways of Development".Developmental biology(6th ed.). Sunderland, Mass: Sinauer Associates.ISBN0-87893-243-7.
- ^Held, Lewis I.(February 2017).Deep Homology?: Uncanny Similarities of Humans and Flies Uncovered by Evo-Devo.Cambridge University Press. pp. viii and throughout.ISBN978-1316601211.
- ^Zimmer, Carl(2016-08-17)."From Fins Into Hands: Scientists Discover a Deep Evolutionary Link".The New York Times.Retrieved21 October2016.
- ^Scharff, Petri; Constance, Jane (July 2011)."Evo-Devo, Deep Homology and FoxP2: Implications for the Evolution of Speech and Language".Philos. Trans. R. Soc. B.366(1574): 2124–2140.doi:10.1098/rstb.2011.0001.PMC3130369.PMID21690130.
- ^"Department of Cancer Biology - Cancer Stem Cells".Mayo Clinic.Retrieved2023-04-10.
- ^abcNiculescu, Vladimir F. (April 4, 2022)."Cancer genes and cancer stem cells in tumorigenesis: Evolutionary deep homology and controversies".Genes & Diseases.9(5): 1234–1247.doi:10.1016/j.gendis.2022.03.010.PMC9293697.PMID35873035.
- ^Zimmer, Carl(April 26, 2010)."The Search for Genes Leads to Unexpected Places".The New York Times.
- ^McGary, K. L.; Park, T. J.; Woods, J. O.; Cha, H. J.; Wallingford, J. B.;Marcotte, E. M.(April 2010)."Systematic discovery of nonobvious human disease models through orthologous phenotypes"(PDF).Proceedings of the National Academy of Sciences.107(14): 6544–9.Bibcode:2010PNAS..107.6544M.doi:10.1073/pnas.0910200107.PMC2851946.PMID20308572.
