Enoyl CoA isomerase
| Δ3-Δ2-Enoyl-CoA isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 3,2-trans-enoyl-CoA isomerase trimer, Human | |||||||||
| Identifiers | |||||||||
| EC no. | 5.3.3.8 | ||||||||
| CAS no. | 62213-29-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDBstructures | RCSB PDBPDBePDBsum | ||||||||
| Gene Ontology | AmiGO/QuickGO | ||||||||
| |||||||||
Enoyl-CoA-(∆) isomerase(EC5.3.3.8,also known asdodecenoyl-CoA-(∆) isomerase,3,2-trans-enoyl-CoA isomerase,∆3(cis),∆2(trans)-enoyl-CoA isomerase,oracetylene-allene isomerase,[1]is anenzymethatcatalyzesthe conversion ofcis- or trans-double bondsofcoenzyme A(CoA) boundfatty acidsat gamma-carbon(position 3) to transdouble bondsat beta-carbon(position 2) as below:
This enzyme has an important role in themetabolismofunsaturated fatty acidsinbeta oxidation.
Mechanism
[edit]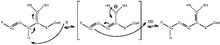
Enoyl-CoAisomeraseis involved in thebeta-oxidation,one of the most frequently used pathways infatty acid degradation,ofunsaturated fatty acidswithdouble bondsat odd-numberedcarbonpositions.[2]It does so by shifting the position of thedouble bondsin the acyl-CoAintermediatesand converting 3-cis or trans-enoyl-CoA to 2-trans-enoyl-CoA. Since the key step in the degradation offatty acidswithdouble bondsat even-numberedcarbonpositions also produces 3-trans-enoyl-CoA inmammalsandyeasts,enoyl-CoAisomeraseis technically required for theirmetabolismas well.[3]Thereaction mechanismis detailed in figure 1,[4]and thebasethat initiates theisomerizationand NH groups that stabilize theintermediateare located on theactive siteof enoyl-coAisomerase.
As it functions in the step immediately preceding the actualbeta-oxidationand forms adouble bondextending from the beta-carbon(position 2), enoyl-CoAisomeraseis involved in both theNADPH-dependent andNADPH-independent pathways ofbeta-oxidation.[5]Thedouble bondserves as the target ofoxidationandcarbon-to-carbonbond cleavage,thereby shortening thefatty acidchain.
Sub-classification
[edit]Enoyl-CoAisomerasescan be categorized into three classes:
- monofunctionalmitochondrial
- monofunctionalperoxisomal
- multifunctional
The monofunctionalmitochondrialandperoxisomalenzymesare found in themitochondriaandperoxisomesofeukaryotes,respectively. The multifunctionalenzymesare found inbacteriaand in theperoxisomesof someeukaryotes,but they serve two functions: theN-terminaldomain works the same as the other classes of enoyl-CoAisomerasesand theC-terminaldomain works as adehydrogenase,specifically, to 3-hydroxyactyl-CoA.[4]There are two divisions among themitochondrialenoyl Co-A isomerase: short-chain and long-chain [4].[6]In an immunoblot, antibodies were run against all enoyl CoA isomerase. However, two of theseisomeraseshadantibodyattachment: the short chain isomerase and the peroxisomal multifunctional enzyme.[6]There was oneenzymewhich did not have binding specificity to thisantibody:mitochondrial long-chain isomerase. Long-chain isomerase was found when it eluted at a lower potassium phosphateconcentrationin the gradient.[6][7]Thus, the discovery of three sub-classes of enoyl CoA isomerase was made.
Although all three classes ofenzymeshave the same function, there is little overlap among theiramino acidsequences. For example, only 40 out of 302amino acidsequences (13%) are the same between monofunctionalperoxisomalandmitochondrialenzymesinhumans.[4]In fact, inmammals,theperoxisomalenzymehas an extraN-terminaldomain that is not present in themitochondrialcounterpart.[8]Also, it has been found to be asubunitof the peroxisomal trifunctionalenzyme(pTFE) and contributes only to minor cleavages of thefatty acidchain. In that sense, for many higher organisms, themitochondrialenzymeis essential for deriving maximumenergyfromlipidsand fuelingmuscles.[9]

Mitochondria(both short- and long-chain) of ratlivercontain more than one enoyl Co-A isomerase.[10]To further support the idea that short- and long-chain isomerases elute at different concentration of potassium phosphate concentration, they do not share similar primary polypeptide structure, hence they must not be evolutionarily related.[6][11]Peroxisomesofplantsand of ratliverare very different in the way they operate. Despite theirprimary structuresimilarities, there are differences among the different specimen. To begin with, theperoxisomesof ratliverare a multifunctionalenzymeincluding enoyl-CoA isomerase, enoyl-CoAhydratase,and L-(−)-3-hydroxyacyl-CoAdehydrogenase.[12]Three differentenzymesreside on this entity (multifunctional protein) allowing thisenzymeto perform isomerization, hydration, and dehydration.[13][14]Isomerase activity on the multifunctionalenzymeoccurs at the amino-terminalcatalytichalf of the protein along with thehydrataseactivity.[15]Thedehydrogenaseactivity of enoyl-CoA occurs in the carboxyl-terminal.[15]Upon further investigation of the CoAbinding siteon the amino-terminal half of the multifunctionalprotein,the CoAsubstrateis not transferred through the aqueous phase from the isomerization phase to the site of hydration or does not have a bulk phase.[11][16]This removes the need for a substrate transferringenzyme.[17]On the other hand, thecotyledonsconvert long-chain 3-trans-enoyl-CoA, long-chain 3-cis-enoyl-CoA, and short-chain 3-cis-enoyl-CoA species into their 2- trans-enoyl-CoA respective forms.[13]As previously mentioned,plantenoyl-CoA isomerase exclusively forms the 2-transisomerasproduct.It does not act on 4-cis-enoyl-CoA species or 2-trans- 4-trans-dienoyl-CoA species.[13]In comparing the products of the plantperoxisomeand the multifunctional enzyme of ratliver,the plant has nohydrataseactivity.[13]The Plant form did not form a 2-cis-isomer (from enoyl-CoA hydratase) or D- or L- 3 hydroxy derivative (L-(−)-3-hydroxyacyl-CoA dehydrogenase): products of multifunctional enzyme of ratliver.[13]The turnover rates of these the two sub divisions ofperoxisomesare very different. The Kcat/Km ratio incotyledonsis 10^6 M-1s-1 which outperforms the ratio.07 * 10^6 M-1s-1.[13]Due to a high turnover rate, the plantperoxisomescontain a lesser amount of enoyl-CoA isomerase than their counterparts in the ratliver.[13]
In rat liver,mitochondrialenoyl CoA isomerase and peroxisomal enoyl CoA isomerase embedded in the multifunctional enzyme have similarities in the primary structure sequence.[15]When comparing the amino-terminal half ofE. coliagainst the amino-terminal half of ratliver,there were primary and secondary structure similarities towards the middle of the amino-terminal end.[15]This conserved region must be important for structure and function of this specific enzyme since showing equally in bothE. coliand ratliver.[15][18]
Structure
[edit]All classes of enoyl-CoAisomerasesbelong to a family ofenzymes,thehydratase/isomeraseorcrotonasesuperfamily, and when examined withx-ray crystallography,exhibit a common structural feature of the family, theN-terminalcore with a spiral fold composed of four turns, each turn consisting of twobeta-sheetsand oneAlpha -helix.[19]
In enoyl-CoAisomerase,the twobeta-sheetsare part of thecatalytic site,since the NH groups ofresiduesfollowing thebeta-sheetsattach to thecarbonyloxygenof the acyl-CoAintermediate.The formation of thisoxyanion holestabilizes thetransition stateof theenzyme-catalyzed reaction.[4]

Moreover, aglutamateresiduelocated next tobody cavitiesfilled with water molecules and lined withhydrophobicorapolarside chainshas also been identified as a part of thecatalytic site.In itsdeprotonatedform, theglutamatecan act as abaseand remove aprotonfrom the acyl-CoAintermediate.Thebody cavitiesaid in rearranging theglutamateside chainto retain theprotonand later deliver it back to the acyl-CoA, on a differentcarbonposition.[4]

The NH-containingresidueshave been identified as Ala70 and Leu126 and theglutamateas Glu158 inperoxisomalenzymesin ayeastspecies,Saccharomyces cerevisiae.Their relative locations on the enzyme can be compared in figure 2.[4]
Theenzymesof thehydratase/isomeraseorcrotonasesuperfamily are typicallytrimericdisksdimerizedintohexamers.The wide range of theirsubstrate-enzymespecificity derives from the variations in the distances between thetrimericdisks and their orientation.[20]However, thehumanmitochondrialenoyl-CoAisomeraseis atrimerand orients thefatty acidtail in a completely different direction from that seen in thehexamers.[8]Thetrimericdisk ofperoxisomalenzymesinSaccharomyces cerevisiaeis displayed in figure 3.[20]
History
[edit]Enoyl-CoAisomerasewas first identified and purified fromratlivermitochondriain the 1960s and 1970s viagel filtrationandion exchange chromatography.[21]Since then, all classes of enoyl-CoAisomerase,mitochondrial,peroxisomaland multifunctional, have been identified in different organisms, including moremammals,plants,andunicellularorganisms.
By 1994, using theratenoyl-CoAisomerasecDNAas ahybridization probe,humanenoyl-CoAisomerasecDNAcould besequencedandcloned.[2]In the same year, the protein itself was isolated, not byaffinitytoratantibodyorcDNAprobes,[3]but by copurificationwith atransferase,human glutathione S-transferases.[22]
In the attempts to examine thehumanenoyl-CoAisomerasein detail, themitochondrialenzymein themammalianliver was identified as a potentialbiological markerformetabolic diseasesdue to its elevated levels in defectivecells,and linked defects infatty acidbeta-oxidationtohumandiseases,[22]to be specified in the next section.
Clinical significance
[edit]Inhumans,defects in thebeta-oxidationmechanism result in hypoketotichyperglycemia,asymptomofstarvation,due to the inefficient utilization offatty acidsas a primary source ofenergy.[9]Themetabolic diseasewas found to be on ageneticlevel:ratswithout thegenesfor enoyl-CoAisomerasealso displayed highblood glucose level.Moreover, abiological markerfor this condition may have been identified as theurineof theseratsincluded high concentrations of medium chainunsaturateddicarboxylic acids,a condition called dicarboxylic aciduria.[9]
More recent studies linkhepatitis C virus(HCV) infection to defects infatty acid degradation,specifically, to that in enoyl-CoAisomerase.[23]HCVis the leading cause ofchronic hepatitis,cirrhosis,andliver cancer,and more than 180 million people are affected globally.[24]Due to the prolongedlatencyof thevirusand no existing cures to rid thevirusspecifically,[25]HCVis a serious problem that is causing more deaths thanHIV/AIDSin theUnited States,[26]but its threat still do not receive adequate attention. The need for aHCV-specific treatment is essential, and according to John Ward, the director of theCDCHepatitis Division, it can save up to 120,000 lives.[26]
According toproteinprofiling in thehumanliverbiopsiesofHCVpatients, a correlation was initially discovered between dysfunctionalmitochondrialprocesses, which includebeta-oxidation,andHCV.[27]As a matter of fact,lipidsplay an important role in thereplicationcycle ofHCV,and in the "in vivo"samples fromHCVpatients, manylipidswere found in abundance to aidHCVinvirusuptake,RNA replication,and secretion fromhost cells.Enzymesthat regulatefatty acid metabolism,including enoyl-CoAisomerase,were also similarlyupregulated.[23]Gene silencingtechniques revealed that enoyl-CoAisomeraseis essential inHCVRNA replication,and opened ways to stopHCVinfection on anintracellularlevel.[23]
See also
[edit]References
[edit]- ^"ENZYME entry 5.3.3.8".Retrieved1 March2012.
- ^abJanssen U, Fink T, Lichter P, Stoffel W (September 1994). "Human mitochondrial 3,2-trans-enoyl-CoA isomerase (DCI): gene structure and localization to chromosome 16p13.3".Genomics.23(1): 223–8.doi:10.1006/geno.1994.1480.PMID7829074.
- ^abKilponen JM, Häyrinen HM, Rehn M, Hiltunen JK (May 1994)."cDNA cloning and amino acid sequence of human mitochondrial delta 3 delta 2-enoyl-CoA isomerase: comparison of the human enzyme with its rat counterpart, mitochondrial short-chain isomerase".Biochemical Journal.300(1): 1–5.doi:10.1042/bj3000001.PMC1138113.PMID8198519.
- ^abcdefMursula AM, van Aalten DM, Hiltunen JK, Wierenga RK (June 2001). "The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase".J. Mol. Biol.309(4): 845–53.doi:10.1006/jmbi.2001.4671.PMID11399063.S2CID69172923.
- ^Luo MJ, Smeland TE, Shoukry K, Schulz H (January 1994)."Delta 3,5, delta 2,4-dienoyl-CoA isomerase from rat liver mitochondria. Purification and characterization of a new enzyme involved in the beta-oxidation of unsaturated fatty acids".J. Biol. Chem.269(4): 2384–8.doi:10.1016/S0021-9258(17)41957-0.PMID8300563.
- ^abcdKilponen, J. M.; Palosaari, P. M.; Hiltunen, J. K. (1990)."Occurrence of a long-chain delta 3,delta 2-enoyl-CoA isomerase in rat liver".Biochemical Journal.269(1): 223–226.doi:10.1042/bj2690223.PMC1131556.PMID2375752.
- ^Brian V. Geisbrecht; Dai Zhu; Kerstin Schulz; Katja Nau; James C. Morrell; Michael Geraghty; Horst Schulz; Ralf Erdmann; Stephen J. Gould (1998)."Molecular Characterization of Saccharomyces cerevisiae delta3, delta2-Enoyl-CoA Isomerase".Journal of Biological Chemistry.273(50): 33184–33191.doi:10.1074/jbc.273.50.33184.PMID9837886.
- ^abPartanen ST, Novikov DK, Popov AN, Mursula AM, Hiltunen JK, Wierenga RK (September 2004). "The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group".J. Mol. Biol.342(4): 1197–208.doi:10.1016/j.jmb.2004.07.039.PMID15351645.
- ^abcJanssen U, Stoffel W (May 2002)."Disruption of mitochondrial beta -oxidation of unsaturated fatty acids in the 3,2-trans-enoyl-CoA isomerase-deficient mouse".J. Biol. Chem.277(22): 19579–84.doi:10.1074/jbc.M110993200.PMID11916962.
- ^abPalosaari P.M.; Hiltunen, J. K. (1991)."Purification and characterization of a plant peroxisomal delta2,delta3-enoyl-CoA isomerase acting on 3-cis-enoyl-CoA and 3-trans-enoyl-CoA"(PDF).Eur. J. Biochem.196(3): 699–705.doi:10.1111/j.1432-1033.1991.tb15868.x.PMID2013292.
- ^abPtiivi M. Palosaari; Johanna M. Kilponen; Raija T. Sormunenn; Ilmo E. Hassine; J. Kalervo Hiltunen (1989)."Characterization of the Mitochondrial Isonzyme in the Rat"(PDF).Journal of Biological Chemistry.265(6): 3347–3353.PMID2154476.
- ^Gerhard Muller-Newen; Uwe Janssen; Wilhelm Stoffel (1995)."Enoyl-CoA hydratase and isomerase form a superfamily with a common active site glutamate residue".Eur. J. Biochem.228(1): 68–73.doi:10.1111/j.1432-1033.1995.0068o.x.PMID7883013.
- ^abcdefgPalosaari PM, Kilponen JM, Sormunen RT, Hassinen E, Hiltunen JK (1990)."Delta 3,delta 2-enoyl-CoA isomerases. Characterization of the mitochondrial isoenzyme in the rat".J. Biol. Chem.265(6): 3347–53.doi:10.1016/S0021-9258(19)39773-X.PMID2154476.
- ^Dongyan Zhang; Wenfeng Yu; Brian V. Geisbrecht; Stephen J. Gould; Howard Sprecher; Horst Schulz (2002)."Functional Characterization of delta3,delta2-Enoyl-CoA Isomerases from Rat Liver".Journal of Biological Chemistry.277(11): 9127–9132.doi:10.1074/jbc.m112228200.PMID11781327.
- ^abcdePaivi M. Palosaari; Mauno Vihinen; Pekka 1. Mantsalag; Stefan E.H. Alexsonll; Taina Pihlajaniemi; J. Kalervo Hiltunen (1991)."Amino Acid Sequence Similarities of the Mitochondrial Short Chain delta3,delta2-Enoyl-CoA Isomerase and Peroxisomal Multifunctional delta3,delta2- Enoyl-CoA Isomerase, 2-Enoyl-CoA Hydratase, 3-Hydroxyacyl-CoA Dehydrogenase Enzyme in Rat Liver"(PDF).Journal of Biological Chemistry.266(17): 10750–10753.doi:10.1016/S0021-9258(18)99081-2.PMID2040594.
{{cite journal}}:CS1 maint: numeric names: authors list (link) - ^Patricia C. Babbitt; George L. Kenyon (1992). "Ancestry of the 4-Chlorobenzoate Dehalogenase: Analysis of Amino Acid Sequence Identities among Families of Acyl: Adenyl Ligases, Enoyl-CoA Hydratases/Isomerases, and Acyl-CoA Thioesterases".Biochemistry.31(24): 5594–5604.doi:10.1021/bi00139a024.PMID1351742.
- ^Anu M. Mursula; Daan M. F. van Aalten; J. Kalervo Hiltunen; Rik K. Wierenga (2001). "The Crystal Structure of delta3-delta2-Enoyl-CoA Isomerase".Molecular Biology.309(4): 845–853.doi:10.1006/jmbi.2001.4671.PMID11399063.S2CID69172923.
- ^Aner Gurvitz; Anu M. Mursula; Andreas Firzinger; Barbara Hamilton; Seppo H. Kilpela ̈ inen; Andreas Hartig; Helmut Ruis; J. Kalervo Hiltunen; Hanspeter Rottensteiner (1998)."Peroxisomal delta3-cis-delta2-trans-Enoyl-CoA Isomerase Encoded by ECI1 Is Required for Growth of the Yeast Saccharomyces cerevisiae on Unsaturated Fatty Acids".Journal of Biological Chemistry.273(47): 31366–31374.doi:10.1074/jbc.273.47.31366.PMID9813046.
- ^Gurvitz A, Mursula AM, Firzinger A, et al. (November 1998)."Peroxisomal Delta3-cis-Delta2-trans-enoyl-CoA isomerase encoded by ECI1 is required for growth of the yeast Saccharomyces cerevisiae on unsaturated fatty acids".J. Biol. Chem.273(47): 31366–74.doi:10.1074/jbc.273.47.31366.PMID9813046.
- ^abMursula AM, Hiltunen JK, Wierenga RK (January 2004)."Structural studies on delta(3)-delta(2)-enoyl-CoA isomerase: the variable mode of assembly of the trimeric disks of the crotonase superfamily".FEBS Lett.557(1–3): 81–7.doi:10.1016/S0014-5793(03)01450-9.PMID14741345.
- ^Stoffel W, Grol M (December 1978). "Purification and properties of 3-cis-2-trans-enoyl-CoA isomerase (dodecenoyl-CoA delta-isomerase) from rat liver mitochondria".Hoppe-Seyler's Z. Physiol. Chem.359(12): 1777–82.doi:10.1515/bchm2.1978.359.2.1777.PMID738702.
- ^abTakahashi Y, Hirata Y, Burstein Y, Listowsky I (December 1994)."Delta 3, delta 2-enoyl-CoA isomerase is the protein that copurifies with human glutathione S-transferases from S-hexylglutathione affinity matrices".Biochemical Journal.304(3): 849–52.doi:10.1042/bj3040849.PMC1137411.PMID7818490.
- ^abcRasmussen AL, Diamond DL, McDermott JE, et al. (November 2011)."Systems virology identifies a mitochondrial fatty acid oxidation enzyme, dodecenoyl coenzyme A delta isomerase, required for hepatitis C virus replication and likely pathogenesis".J. Virol.85(22): 11646–54.doi:10.1128/JVI.05605-11.PMC3209311.PMID21917952.
- ^Rosen, Hugo R. (June 2011). "Chronic Hepatitis C Infection".The New England Journal of Medicine.364(25): 2429–2438.doi:10.1056/NEJMcp1006613.PMID21696309.S2CID19755395.
- ^Amemiya F, Maekawa S, Itakura Y, et al. (February 2008)."Targeting lipid metabolism in the treatment of hepatitis C virus infection".J. Infect. Dis.197(3): 361–70.doi:10.1086/525287.PMID18248300.
- ^ab"Hepatitis C Kills More Americans Than HIV/AIDS".Voice of America, Health.27 February 2012.Retrieved3 March2012.
- ^Diamond DL, Jacobs JM, Paeper B, et al. (September 2007)."Proteomic profiling of human liver biopsies: hepatitis C virus-induced fibrosis and mitochondrial dysfunction".Hepatology.46(3): 649–57.doi:10.1002/hep.21751.PMID17654742.

