Eukaryote
| Eukaryota Temporal range:Statherian–Present
| |
|---|---|
| Scientific classification | |
| Domain: | Eukaryota (Chatton,1925)Whittaker&Margulis,1978 |
| Subgroups | |
| Synonyms | |
Theeukaryotes(/juːˈkærioʊts,-əts/yoo-KARR-ee-ohts, -əts)[4]constitute thedomainofEukaryaorEukaryota,organismswhosecellshave a membrane-boundnucleus.Allanimals,plants,fungi,and manyunicellular organismsare eukaryotes. They constitute a major group oflife formsalongside the two groups ofprokaryotes:theBacteriaand theArchaea.Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collectiveglobal biomassis much larger than that of prokaryotes.
The eukaryotes seemingly emerged within theAsgard archaea,and are closely related to theHeimdallarchaeia.[5]This implies that there are onlytwo domains of life,Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during thePaleoproterozoic,likely asflagellatedcells. The leadingevolutionarytheory is they were created bysymbiogenesisbetween an anaerobic Asgard archaean and an aerobicproteobacterium,which formed themitochondria.A second episode of symbiogenesis with acyanobacteriumcreated the plants, withchloroplasts.
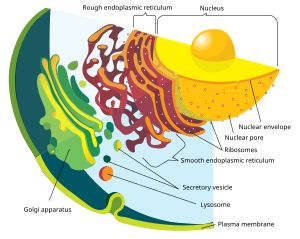
Eukaryotic cells containmembrane-bound organellessuch as thenucleus,theendoplasmic reticulum,and theGolgi apparatus.Eukaryotes may be eitherunicellularormulticellular.In comparison, prokaryotes are typically unicellular. Unicellular eukaryotes are sometimes calledprotists.Eukaryotes can reproduce bothasexuallythroughmitosisandsexuallythroughmeiosisandgametefusion (fertilization).
Diversity
Eukaryotes areorganismsthat range from microscopic singlecells,such aspicozoansunder 3 micrometres across,[6]toanimalslike theblue whale,weighing up to 190tonnesand measuring up to 33.6 metres (110 ft) long,[7]orplantslike thecoast redwood,up to 120 metres (390 ft) tall.[8]Many eukaryotes are unicellular; the informal grouping calledprotistsincludes many of these, with some multicellular forms like thegiant kelpup to 200 feet (61 m) long.[9]The multicellular eukaryotes include the animals, plants, andfungi,but again, these groups too contain many unicellularspecies.[10]Eukaryotic cells are typically much larger than those ofprokaryotes—thebacteriaand thearchaea—having a volume of around 10,000 times greater.[11][12]Eukaryotes represent a small minority of the number oforganisms,but, as many of them are much larger, their collective globalbiomass(468 gigatons) is far larger than that of prokaryotes (77 gigatons), with plants alone accounting for over 81% of the total biomass ofEarth.[13]
- Eukaryotes range in size from single cells to organisms weighing many tons
The eukaryotes are a diverse lineage, consisting mainly ofmicroscopic organisms.[14]Multicellularity in some form hasevolved independentlyat least 25 times within the eukaryotes.[15][16]Complex multicellular organisms, not counting the aggregation ofamoebaeto formslime molds,have evolved within only six eukaryotic lineages:animals,symbiomycotan fungi,brown algae,red algae,green algae,andland plants.[17]Eukaryotes are grouped by genomic similarities, so that groups often lack visible shared characteristics.[14]
Distinguishing features
Nucleus
The defining feature of eukaryotes is thattheir cellshavenuclei.This gives them their name, from theGreekεὖ(eu,"well" or "good" ) andκάρυον(karyon,"nut" or "kernel", here meaning "nucleus" ).[18]Eukaryotic cells have a variety of internal membrane-bound structures, calledorganelles,and acytoskeletonwhich defines the cell's organization and shape. The nucleus stores the cell'sDNA,which is divided into linear bundles calledchromosomes;[19]these are separated into two matching sets by amicrotubular spindleduring nuclear division, in the distinctively eukaryotic process ofmitosis.[20]
Biochemistry
Eukaryotes differ from prokaryotes in multiple ways, with unique biochemical pathways such assteranesynthesis.[21]The eukaryotic signatureproteinshave no homology to proteins in other domains of life, but appear to be universal among eukaryotes. They include the proteins of the cytoskeleton, the complextranscriptionmachinery, the membrane-sorting systems, thenuclear pore,and someenzymesin the biochemical pathways.[22]
Internal membranes
Eukaryote cells include a variety of membrane-bound structures, together forming theendomembrane system.[23]Simple compartments, calledvesiclesandvacuoles,can form by budding off other membranes. Many cells ingest food and other materials through a process ofendocytosis,where the outer membraneinvaginatesand then pinches off to form a vesicle.[24]Some cell products can leave in a vesicle throughexocytosis.[25]
The nucleus is surrounded by a double membrane known as thenuclear envelope,withnuclear poresthat allow material to move in and out.[26]Various tube- and sheet-like extensions of the nuclear membrane form theendoplasmic reticulum,which is involved inprotein transportand maturation. It includes the rough endoplasmic reticulum, covered inribosomeswhich synthesize proteins; these enter the interior space or lumen. Subsequently, they generally enter vesicles, which bud off from the smooth endoplasmic reticulum.[27]In most eukaryotes, these protein-carrying vesicles are released and further modified in stacks of flattened vesicles (cisternae), theGolgi apparatus.[28]
Vesicles may be specialized; for instance,lysosomescontaindigestive enzymesthat break downbiomoleculesin the cytoplasm.[29]
Mitochondria

Mitochondriaare organelles in eukaryotic cells. The mitochondrion is commonly called "the powerhouse of the cell",[30]for its function providing energy by oxidising sugars or fats to produce the energy-storing moleculeATP.[31][32]Mitochondria have two surroundingmembranes,each aphospholipid bilayer;theinnerof which is folded into invaginations calledcristaewhereaerobic respirationtakes place.[33]
Mitochondria containtheir own DNA,which has close structural similarities tobacterial DNA,from which it originated, and which encodesrRNAandtRNAgenes that produce RNA which is closer in structure to bacterial RNA than to eukaryote RNA.[34]
Some eukaryotes, such as themetamonadsGiardiaandTrichomonas,and the amoebozoanPelomyxa,appear to lack mitochondria, but all contain mitochondrion-derived organelles, likehydrogenosomesormitosomes,having lost their mitochondria secondarily.[35]They obtain energy by enzymatic action in the cytoplasm.[36][35]
Plastids

Plants and various groups ofalgaehaveplastidsas well as mitochondria. Plastids, like mitochondria, havetheir own DNAand are developed fromendosymbionts,in this casecyanobacteria.They usually take the form ofchloroplastswhich, like cyanobacteria, containchlorophylland produce organic compounds (such asglucose) throughphotosynthesis.Others are involved in storing food. Although plastids probably had a single origin, not all plastid-containing groups are closely related. Instead, some eukaryotes have obtained them from others throughsecondary endosymbiosisor ingestion.[37]The capture and sequestering of photosynthetic cells and chloroplasts,kleptoplasty,occurs in many types of modern eukaryotic organisms.[38][39]
Cytoskeletal structures
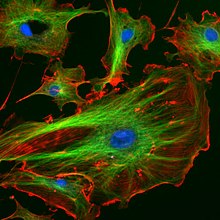
Thecytoskeletonprovides stiffening structure and points of attachment for motor structures that enable the cell to move, change shape, or transport materials. The motor structures aremicrofilamentsofactinandactin-binding proteins,including α-actinin,fimbrin,andfilaminare present in submembranouscortical layersand bundles.Motor proteinsof microtubules,dyneinandkinesin,andmyosinof actin filaments, provide dynamic character of the network.[40][41]
Many eukaryotes have long slender motile cytoplasmic projections, calledflagella,or multiple shorter structures calledcilia.These organellesare variously involved in movement, feeding, and sensation. They are composed mainly oftubulin,and are entirely distinct from prokaryotic flagella. They are supported by a bundle ofmicrotubulesarising from acentriole,characteristically arranged as nine doublets surrounding two singlets. Flagella may have hairs (mastigonemes), as in manyStramenopiles.Their interior is continuous with the cell'scytoplasm.[42][43]
Centrioles are often present, even in cells and groups that do not have flagella, butconifersandflowering plantshave neither. They generally occur in groups that give rise to various microtubular roots. These form a primary component of the cytoskeleton, and are often assembled over the course of several cell divisions, with one flagellum retained from the parent and the other derived from it. Centrioles produce the spindle during nuclear division.[44]
Cell wall
The cells of plants, algae, fungi and mostchromalveolates,but not animals, are surrounded by a cell wall. This is a layer outside thecell membrane,providing the cell with structural support, protection, and a filtering mechanism. The cell wall also preventsover-expansionwhen water enters the cell.[45]
The majorpolysaccharidesmaking up the primary cell wall ofland plantsarecellulose,hemicellulose,andpectin.The cellulosemicrofibrilsare linked together with hemicellulose, embedded in a pectin matrix. The most common hemicellulose in the primary cell wall isxyloglucan.[46]
Sexual reproduction

Eukaryotes have a life cycle that involvessexual reproduction,alternating between ahaploidphase, where only one copy of each chromosome is present in each cell, and adiploidphase, with two copies of each chromosome in each cell. The diploid phase is formed by fusion of two haploid gametes, such aseggsandspermatozoa,to form azygote;this may grow into a body, with its cells dividing bymitosis,and at some stage produce haploid gametes throughmeiosis,a division that reduces the number of chromosomes and createsgenetic variability.[47]There is considerable variation in this pattern. Plants have bothhaploid and diploid multicellular phases.[48]Eukaryotes have lower metabolic rates and longer generation times than prokaryotes, because they are larger and therefore have a smaller surface area to volume ratio.[49]
Theevolution of sexual reproductionmay be a primordial characteristic of eukaryotes. Based on a phylogenetic analysis, Dacks andRogerhave proposed that facultative sex was present in the group's common ancestor.[50]A core set of genes that function in meiosis is present in bothTrichomonas vaginalisandGiardia intestinalis,two organisms previously thought to be asexual.[51][52]Since these two species are descendants of lineages that diverged early from the eukaryotic evolutionary tree, core meiotic genes, and hence sex, were likely present in the common ancestor of eukaryotes.[51][52]Species once thought to be asexual, such asLeishmaniaparasites, have a sexual cycle.[53]Amoebae, previously regarded as asexual, may be anciently sexual; while present-day asexual groups could have arisen recently.[54]
Evolution
History of classification
Inantiquity,the two lineages ofanimalsandplantswere recognized byAristotleandTheophrastus.The lineages were given thetaxonomic rankofKingdombyLinnaeusin the 18th century. Though he included thefungiwith plants with some reservations, it was later realized that they are quite distinct and warrant a separate kingdom.[56]The various single-cell eukaryotes were originally placed with plants or animals when they became known. In 1818, the German biologistGeorg A. Goldfusscoined the wordprotozoato refer to organisms such asciliates,[57]and this group was expanded untilErnst Haeckelmade it a kingdom encompassing all single-celled eukaryotes, theProtista,in 1866.[58][59][60]The eukaryotes thus came to be seen as four kingdoms:
The protists were at that time thought to be "primitive forms", and thus anevolutionary grade,united by their primitive unicellular nature.[59]Understanding of the oldest branchings in thetree of lifeonly developed substantially withDNA sequencing,leading to a system ofdomainsrather than kingdoms as top level rank being put forward byCarl Woese,Otto Kandler,andMark Wheelisin 1990, uniting all the eukaryote kingdoms in the domain "Eucarya", stating, however, that"'eukaryotes' will continue to be an acceptable common synonym ".[2][61]In 1996, the evolutionary biologistLynn Margulisproposed to replace Kingdoms and Domains with "inclusive" names to create a "symbiosis-based phylogeny", giving the description "Eukarya (symbiosis-derived nucleated organisms)".[3]
Phylogeny
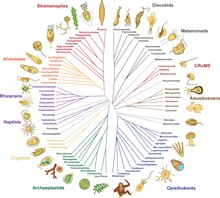
By 2014, a rough consensus started to emerge from the phylogenomic studies of the previous two decades.[10][62]The majority of eukaryotes can be placed in one of two large clades dubbedAmorphea(similar in composition to theunikonthypothesis) and theDiphoda(formerly bikonts), which includes plants and most algal lineages. A third major grouping, theExcavata,has been abandoned as a formal group as it isparaphyletic.[63]The proposed phylogeny below includes only one group of excavates (Discoba),[64]and incorporates the 2021 proposal thatpicozoansare close relatives of rhodophytes.[65]TheProvoraare a group of microbial predators discovered in 2022.[1]
| Eukaryotes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2200 mya |
One view of the great kingdoms and their stem groups.[64][66][67][14]TheMetamonadaare hard to place, being sister possibly toDiscobaor toMalawimonada[14]or being a paraphyletic group external toall other eukaryotes.[68]
Origin of eukaryotes
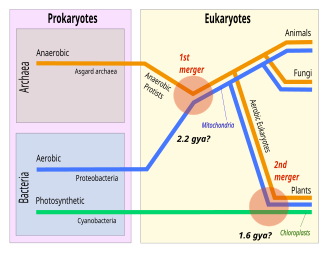
The origin of the eukaryotic cell, oreukaryogenesis,is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. Thelast eukaryotic common ancestor(LECA) is the hypothetical origin of all living eukaryotes,[70]and was most likely abiological population,not a single individual.[71]The LECA is believed to have been a protist with a nucleus, at least onecentrioleandflagellum,facultatively aerobic mitochondria, sex (meiosisandsyngamy), a dormantcystwith a cell wall ofchitinorcellulose,andperoxisomes.[72][73][74]
Anendosymbiotic unionbetween a motileanaerobicarchaean and an aerobicAlpha proteobacteriumgave rise to the LECA and all eukaryotes, withmitochondria.A second, much later endosymbiosis with a cyanobacterium gave rise to the ancestor of plants, withchloroplasts.[69]
The presence of eukaryotic biomarkers in archaea points towards an archaeal origin. The genomes ofAsgardarchaea have plenty ofEukaryotic signature proteingenes, which play a crucial role in the development of thecytoskeletonand complex cellular structures characteristic of eukaryotes. In 2022,cryo-electron tomographydemonstrated that Asgard archaea have a complexactin-based cytoskeleton, providing the first direct visual evidence of the archaeal ancestry of eukaryotes.[75]
Fossils
The timing of the origin of eukaryotes is hard to determine but the discovery ofQingshania magnificia,the earliest multicelluar eukaryote from North China which lived during 1.635 billion years ago, suggests that the crown group eukaryotes would have originated from the latePaleoproterozoic(Statherian); the earliest unequivocal unicellular eukaryotes which lived during approximately 1.65 billion years ago are also discovered from North China:Tappania plana,Shuiyousphaeridium macroreticulatum,Dictyosphaera macroreticulata,Germinosphaera alveolata,andValeria lophostriata.[76]
Someacritarchsare known from at least 1.65 billion years ago, and a fossil,Grypania,which may be an alga, is as much as 2.1 billion years old.[77][78]The"problematic"[79]fossilDiskagmahas been found inpaleosols2.2 billion years old.[79]

Structures proposed to represent "large colonial organisms" have been found in theblack shalesof thePalaeoproterozoicsuch as theFrancevillian B Formation,inGabon,dubbed the "Francevillian biota"which is dated at 2.1 billion years old.[80][81]However, the status of these structures as fossils is contested, with other authors suggesting that they might representpseudofossils.[82]The oldest fossils than can unambiguously be assigned to eukaryotes are from the Ruyang Group of China, dating to approximately 1.8-1.6 billion years ago.[83]Fossils that are clearly related to modern groups start appearing an estimated 1.2 billion years ago, in the form ofred algae,though recent work suggests the existence of fossilizedfilamentous algaein theVindhyabasin dating back perhaps to 1.6 to 1.7 billion years ago.[84]
The presence ofsteranes,eukaryotic-specificbiomarkers,inAustralianshalespreviously indicated that eukaryotes were present in these rocks dated at 2.7 billion years old,[21][85]but these Archaean biomarkers have been rebutted as later contaminants.[86]The oldest valid biomarker records are only around 800 million years old.[87]In contrast, a molecular clock analysis suggests the emergence of sterol biosynthesis as early as 2.3 billion years ago.[88]The nature of steranes as eukaryotic biomarkers is further complicated by the production ofsterolsby some bacteria.[89][90]
Whenever their origins, eukaryotes may not have become ecologically dominant until much later; a massive increase in thezinc compositionof marine sediments800million years agohas been attributed to the rise of substantial populations of eukaryotes, which preferentially consume and incorporatezincrelative to prokaryotes, approximately a billion years after their origin (at the latest).[91]
See also
- Eukaryote hybrid genome
- List of sequenced eukaryotic genomes
- Parakaryon myojinensis
- Vault (organelle)
References
- ^abTikhonenkov DV, Mikhailov KV, Gawryluk RM, et al. (December 2022). "Microbial predators form a new supergroup of eukaryotes".Nature.612(7941): 714–719.Bibcode:2022Natur.612..714T.doi:10.1038/s41586-022-05511-5.PMID36477531.S2CID254436650.
- ^abWoese CR,Kandler O,Wheelis ML(June 1990)."Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya".Proceedings of the National Academy of Sciences of the United States of America.87(12): 4576–4579.Bibcode:1990PNAS...87.4576W.doi:10.1073/pnas.87.12.4576.PMC54159.PMID2112744.
- ^abMargulis L(6 February 1996)."Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life".Proceedings of the National Academy of Sciences.93(3): 1071–1076.Bibcode:1996PNAS...93.1071M.doi:10.1073/pnas.93.3.1071.PMC40032.PMID8577716.
- ^"eukaryote".Merriam-Webster Dictionary.Merriam-Webster.
- ^Eme, Laura; Tamarit, Daniel; Caceres, Eva F.; Stairs, Courtney W.; De Anda, Valerie; Schön, Max E.; Seitz, Kiley W.; Dombrowski, Nina; Lewis, William H.; Homa, Felix; Saw, Jimmy H.; Lombard, Jonathan; Nunoura, Takuro; Li, Wen-Jun; Hua, Zheng-Shuang; Chen, Lin-Xing; Banfield, Jillian F.; John, Emily St; Reysenbach, Anna-Louise; Stott, Matthew B.; Schramm, Andreas; Kjeldsen, Kasper U.; Teske, Andreas P.; Baker, Brett J.; Ettema, Thijs J. G. (29 June 2023)."Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes".Nature.618(7967): 992–999.Bibcode:2023Natur.618..992E.doi:10.1038/s41586-023-06186-2.ISSN1476-4687.PMC10307638.PMID37316666.
- ^Seenivasan R, Sausen N, Medlin LK, Melkonian M (26 March 2013)."Picomonas judraskeda Gen. Et Sp. Nov.: The First Identified Member of the Picozoa Phylum Nov., a Widespread Group of Picoeukaryotes, Formerly Known as 'Picobiliphytes'".PLOS ONE.8(3): e59565.Bibcode:2013PLoSO...859565S.doi:10.1371/journal.pone.0059565.PMC3608682.PMID23555709.
- ^Wood G (1983).The Guinness Book of Animal Facts and Feats.Enfield, Middlesex: Guinness Superlatives.ISBN978-0-85112-235-9.
- ^Earle CJ, ed. (2017)."Sequoia sempervirens".The Gymnosperm Database.Archivedfrom the original on 1 April 2016.Retrieved15 September2017.
- ^van den Hoek C, Mann D, Jahns H (1995).Algae An Introduction to Phycology.Cambridge: Cambridge University Press.ISBN0-521-30419-9.Archivedfrom the original on 10 February 2023.Retrieved7 April2023.
- ^abBurki F (May 2014)."The eukaryotic tree of life from a global phylogenomic perspective".Cold Spring Harbor Perspectives in Biology.6(5): a016147.doi:10.1101/cshperspect.a016147.PMC3996474.PMID24789819.
- ^DeRennaux B (2001). "Eukaryotes, Origin of".Encyclopedia of Biodiversity.Vol. 2. Elsevier. pp. 329–332.doi:10.1016/b978-0-12-384719-5.00174-x.ISBN9780123847201.
- ^Yamaguchi M, Worman CO (2014)."Deep-sea microorganisms and the origin of the eukaryotic cell"(PDF).Japanese Journal of Protozoology.47(1, 2): 29–48. Archived fromthe original(PDF)on 9 August 2017.
- ^Bar-On, Yinon M.; Phillips, Rob; Milo, Ron (17 May 2018)."The biomass distribution on Earth".Proceedings of the National Academy of Sciences.115(25): 6506–6511.Bibcode:2018PNAS..115.6506B.doi:10.1073/pnas.1711842115.ISSN0027-8424.PMC6016768.PMID29784790.
- ^abcdBurki F, Roger AJ, Brown MW, Simpson AG (2020)."The New Tree of Eukaryotes".Trends in Ecology & Evolution.35(1). Elsevier BV: 43–55.Bibcode:2020TEcoE..35...43B.doi:10.1016/j.tree.2019.08.008.ISSN0169-5347.PMID31606140.S2CID204545629.Cite error: The named reference "Burki Roger Brown Simpson 2020 pp. 43–55" was defined multiple times with different content (see thehelp page).
- ^Grosberg RK, Strathmann RR (2007)."The evolution of multicellularity: A minor major transition?"(PDF).Annu Rev Ecol Evol Syst.38:621–654.doi:10.1146/annurev.ecolsys.36.102403.114735.Archived(PDF)from the original on 14 March 2023.Retrieved8 April2023.
- ^Parfrey L, Lahr D (2013)."Multicellularity arose several times in the evolution of eukaryotes"(PDF).BioEssays.35(4): 339–347.doi:10.1002/bies.201200143.PMID23315654.S2CID13872783.Archived(PDF)from the original on 25 July 2014.Retrieved8 April2023.
- ^Popper ZA, Michel G, Hervé C, Domozych DS, Willats WG, Tuohy MG, Kloareg B, Stengel DB (2011). "Evolution and diversity of plant cell walls: From algae to flowering plants".Annual Review of Plant Biology.62:567–590.doi:10.1146/annurev-arplant-042110-103809.hdl:10379/6762.PMID21351878.S2CID11961888.
- ^Harper, Douglas."eukaryotic".Online Etymology Dictionary.
- ^Bonev B, Cavalli G (14 October 2016). "Organization and function of the 3D genome".Nature Reviews Genetics.17(11): 661–678.doi:10.1038/nrg.2016.112.hdl:2027.42/151884.PMID27739532.S2CID31259189.
- ^O'Connor, Clare (2008)."Chromosome Segregation: The Role of Centromeres".Nature Education.Retrieved18 February2024.
eukar
- ^abBrocks JJ, Logan GA, Buick R, Summons RE (August 1999). "Archean molecular fossils and the early rise of eukaryotes".Science.285(5430): 1033–1036.Bibcode:1999Sci...285.1033B.CiteSeerX10.1.1.516.9123.doi:10.1126/science.285.5430.1033.PMID10446042.
- ^Hartman H, Fedorov A (February 2002)."The origin of the eukaryotic cell: a genomic investigation".Proceedings of the National Academy of Sciences of the United States of America.99(3): 1420–5.Bibcode:2002PNAS...99.1420H.doi:10.1073/pnas.032658599.PMC122206.PMID11805300.
- ^Linka M, Weber AP (2011)."Evolutionary Integration of Chloroplast Metabolism with the Metabolic Networks of the Cells".In Burnap RL, Vermaas WF (eds.).Functional Genomics and Evolution of Photosynthetic Systems.Springer. p. 215.ISBN978-94-007-1533-2.Archivedfrom the original on 29 May 2016.Retrieved27 October2015.
- ^Marsh M (2001).Endocytosis.Oxford University Press. p. vii.ISBN978-0-19-963851-2.
- ^Stalder D, Gershlick DC (November 2020)."Direct trafficking pathways from the Golgi apparatus to the plasma membrane".Seminars in Cell & Developmental Biology.107:112–125.doi:10.1016/j.semcdb.2020.04.001.PMC7152905.PMID32317144.
- ^Hetzer MW (March 2010)."The nuclear envelope".Cold Spring Harbor Perspectives in Biology.2(3): a000539.doi:10.1101/cshperspect.a000539.PMC2829960.PMID20300205.
- ^"Endoplasmic Reticulum (Rough and Smooth)".British Society for Cell Biology.Archivedfrom the original on 24 March 2019.Retrieved12 November2017.
- ^"Golgi Apparatus".British Society for Cell Biology. Archived fromthe originalon 13 November 2017.Retrieved12 November2017.
- ^"Lysosome".British Society for Cell Biology. Archived fromthe originalon 13 November 2017.Retrieved12 November2017.
- ^Saygin D, Tabib T, Bittar HE, et al. (July 1957)."Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension".Pulmonary Circulation.10(1): 131–144.Bibcode:1957SciAm.197a.131S.doi:10.1038/scientificamerican0757-131.PMC7052475.PMID32166015.
- ^Voet D, Voet JC, Pratt CW (2006).Fundamentals of Biochemistry(2nd ed.). John Wiley and Sons. pp.547, 556.ISBN978-0471214953.
- ^Mack S (1 May 2006)."Re: Are there eukaryotic cells without mitochondria?".madsci.org.Archivedfrom the original on 24 April 2014.Retrieved24 April2014.
- ^Zick M, Rabl R, Reichert AS (January 2009). "Cristae formation-linking ultrastructure and function of mitochondria".Biochimica et Biophysica Acta (BBA) - Molecular Cell Research.1793(1): 5–19.doi:10.1016/j.bbamcr.2008.06.013.PMID18620004.
- ^Watson J, Hopkins N, Roberts J, Steitz JA, Weiner A (1988)."28: The Origins of Life".Molecular Biology of the Gene(Fourth ed.). Menlo Park, California: The Benjamin/Cummings Publishing Company, Inc. p.1154.ISBN978-0-8053-9614-0.
- ^abKarnkowska A, Vacek V, Zubáčová Z, et al. (May 2016)."A Eukaryote without a Mitochondrial Organelle".Current Biology.26(10): 1274–1284.Bibcode:2016CBio...26.1274K.doi:10.1016/j.cub.2016.03.053.PMID27185558.
- ^Davis JL (13 May 2016)."Scientists Shocked To Discover Eukaryote With NO Mitochondria".IFL Science.Archived fromthe originalon 17 February 2019.Retrieved13 May2016.
- ^Sato N (2006). "Origin and Evolution of Plastids: Genomic View on the Unification and Diversity of Plastids". In Wise RR, Hoober JK (eds.).The Structure and Function of Plastids.Advances in Photosynthesis and Respiration. Vol. 23. Springer Netherlands. pp. 75–102.doi:10.1007/978-1-4020-4061-0_4.ISBN978-1-4020-4060-3.
- ^Minnhagen S, Carvalho WF, Salomon PS, Janson S (September 2008). "Chloroplast DNA content in Dinophysis (Dinophyceae) from different cell cycle stages is consistent with kleptoplasty".Environ. Microbiol.10(9): 2411–7.Bibcode:2008EnvMi..10.2411M.doi:10.1111/j.1462-2920.2008.01666.x.PMID18518896.
- ^Bodył A (February 2018). "Did some red alga-derived plastids evolve via kleptoplastidy? A hypothesis".Biological Reviews of the Cambridge Philosophical Society.93(1): 201–222.doi:10.1111/brv.12340.PMID28544184.S2CID24613863.
- ^Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (1 January 2002). "Molecular Motors".Molecular Biology of the Cell(4th ed.). New York: Garland Science.ISBN978-0-8153-3218-3.Archivedfrom the original on 8 March 2019.Retrieved6 April2023.
- ^Sweeney HL, Holzbaur EL (May 2018)."Motor Proteins".Cold Spring Harbor Perspectives in Biology.10(5): a021931.doi:10.1101/cshperspect.a021931.PMC5932582.PMID29716949.
- ^Bardy SL, Ng SY, Jarrell KF (February 2003)."Prokaryotic motility structures".Microbiology.149(Pt 2): 295–304.doi:10.1099/mic.0.25948-0.PMID12624192.
- ^Silflow CD, Lefebvre PA (December 2001)."Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii".Plant Physiology.127(4): 1500–7.doi:10.1104/pp.010807.PMC1540183.PMID11743094.
- ^Vorobjev IA, Nadezhdina ES (1987).The centrosome and its role in the organization of microtubules.International Review of Cytology. Vol. 106. pp. 227–293.doi:10.1016/S0074-7696(08)61714-3.ISBN978-0-12-364506-7.PMID3294718.
- ^Howland JL (2000).The Surprising Archaea: Discovering Another Domain of Life.Oxford: Oxford University Press. pp. 69–71.ISBN978-0-19-511183-5.
- ^Fry SC (1989). "The Structure and Functions of Xyloglucan".Journal of Experimental Botany.40(1): 1–11.doi:10.1093/jxb/40.1.1.
- ^Hamilton MB (2009).Population genetics.Wiley-Blackwell.p.55.ISBN978-1-4051-3277-0.
- ^Taylor TN, Kerp H, Hass H (2005)."Life history biology of early land plants: Deciphering the gametophyte phase".Proceedings of the National Academy of Sciences of the United States of America.102(16): 5892–5897.doi:10.1073/pnas.0501985102.PMC556298.PMID15809414.
- ^Lane N(June 2011)."Energetics and genetics across the prokaryote-eukaryote divide".Biology Direct.6(1): 35.doi:10.1186/1745-6150-6-35.PMC3152533.PMID21714941.
- ^Dacks J, Roger AJ (June 1999). "The first sexual lineage and the relevance of facultative sex".Journal of Molecular Evolution.48(6): 779–783.Bibcode:1999JMolE..48..779D.doi:10.1007/PL00013156.PMID10229582.S2CID9441768.
- ^abRamesh MA, Malik SB, Logsdon JM (January 2005)."A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis".Current Biology.15(2): 185–191.Bibcode:2005CBio...15..185R.doi:10.1016/j.cub.2005.01.003.PMID15668177.S2CID17013247.
- ^abMalik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM (August 2007). Hahn MW (ed.)."An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis".PLOS ONE.3(8): e2879.Bibcode:2008PLoSO...3.2879M.doi:10.1371/journal.pone.0002879.PMC2488364.PMID18663385.
- ^Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL (April 2009)."Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector".Science.324(5924): 265–268.Bibcode:2009Sci...324..265A.doi:10.1126/science.1169464.PMC2729066.PMID19359589.
- ^Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E (July 2011)."The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms".Proceedings: Biological Sciences.278(1715): 2081–2090.doi:10.1098/rspb.2011.0289.PMC3107637.PMID21429931.
- ^Patrick J. Keeling;Yana Eglit (21 November 2023)."Openly available illustrations as tools to describe eukaryotic microbial diversity".PLOS Biology.21(11): e3002395.doi:10.1371/JOURNAL.PBIO.3002395.ISSN1544-9173.PMC10662721.PMID37988341.WikidataQ123558544.
- ^Moore RT (1980). "Taxonomic proposals for the classification of marine yeasts and other yeast-like fungi including the smuts".Botanica Marina.23(6): 361–373.doi:10.1515/bot-1980-230605.
- ^Goldfuß (1818)."Ueber die Classification der Zoophyten"[On the classification of zoophytes].Isis, Oder, Encyclopädische Zeitung von Oken(in German).2(6): 1008–1019.Archivedfrom the original on 24 March 2019.Retrieved15 March2019.From p. 1008:"Erste Klasse. Urthiere. Protozoa."(First class. Primordial animals. Protozoa.) [Note: each column of each page of this journal is numbered; there are two columns per page.]
- ^Scamardella JM (1999)."Not plants or animals: a brief history of the origin of Kingdoms Protozoa, Protista and Protoctista"(PDF).International Microbiology.2(4): 207–221.PMID10943416.Archived fromthe original(PDF)on 14 June 2011.
- ^abRothschild LJ(1989)."Protozoa, Protista, Protoctista: what's in a name?".Journal of the History of Biology.22(2): 277–305.doi:10.1007/BF00139515.PMID11542176.S2CID32462158.Archivedfrom the original on 4 February 2020.Retrieved4 February2020.
- ^Whittaker RH (January 1969). "New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms".Science.163(3863): 150–60.Bibcode:1969Sci...163..150W.CiteSeerX10.1.1.403.5430.doi:10.1126/science.163.3863.150.PMID5762760.
- ^Knoll AH(1992). "The Early Evolution of Eukaryotes: A Geological Perspective".Science.256(5057): 622–627.Bibcode:1992Sci...256..622K.doi:10.1126/science.1585174.PMID1585174.
Eucarya, or eukaryotes
- ^Burki F, Kaplan M, Tikhonenkov DV, et al. (January 2016)."Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista".Proceedings: Biological Sciences.283(1823): 20152802.doi:10.1098/rspb.2015.2802.PMC4795036.PMID26817772.
- ^Adl SM, Bass D, Lane CE, et al. (January 2019)."Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes".The Journal of Eukaryotic Microbiology.66(1): 4–119.doi:10.1111/jeu.12691.PMC6492006.PMID30257078.
- ^abBrown MW, Heiss AA, Kamikawa R, Inagaki Y, Yabuki A, Tice AK, Shiratori T, Ishida KI, Hashimoto T, Simpson A, Roger A (19 January 2018)."Phylogenomics Places Orphan Protistan Lineages in a Novel Eukaryotic Super-Group".Genome Biology and Evolution.10(2): 427–433.doi:10.1093/gbe/evy014.PMC5793813.PMID29360967.
- ^Schön ME, Zlatogursky VV, Singh RP, et al. (2021)."Picozoa are archaeplastids without plastid".Nature Communications.12(1): 6651.bioRxiv10.1101/2021.04.14.439778.doi:10.1038/s41467-021-26918-0.PMC8599508.PMID34789758.S2CID233328713.Archivedfrom the original on 2 February 2024.Retrieved20 December2021.
- ^Schön ME, Zlatogursky VV, Singh RP, et al. (2021)."Picozoa are archaeplastids without plastid".Nature Communications.12(1): 6651.bioRxiv10.1101/2021.04.14.439778.doi:10.1038/s41467-021-26918-0.PMC8599508.PMID34789758.S2CID233328713.
- ^Tikhonenkov DV, Mikhailov KV, Gawryluk RM, et al. (December 2022). "Microbial predators form a new supergroup of eukaryotes".Nature.612(7941): 714–719.doi:10.1038/s41586-022-05511-5.PMID36477531.S2CID254436650.
- ^Al Jewari, Caesar; Baldauf, Sandra L. (28 April 2023)."An excavate root for the eukaryote tree of life".Science Advances.9(17): eade4973.Bibcode:2023SciA....9E4973A.doi:10.1126/sciadv.ade4973.ISSN2375-2548.PMC10146883.PMID37115919.
- ^abLatorre A, Durban A, Moya A, Pereto J (2011)."The role of symbiosis in eukaryotic evolution".In Gargaud M, López-Garcìa P, Martin H (eds.).Origins and Evolution of Life: An astrobiological perspective.Cambridge: Cambridge University Press. pp. 326–339.ISBN978-0-521-76131-4.Archivedfrom the original on 24 March 2019.Retrieved27 August2017.
- ^Gabaldón T (October 2021). "Origin and Early Evolution of the Eukaryotic Cell".Annual Review of Microbiology.75(1): 631–647.doi:10.1146/annurev-micro-090817-062213.PMID34343017.S2CID236916203.
- ^O'Malley MA, Leger MM, Wideman JG, Ruiz-Trillo I (March 2019). "Concepts of the last eukaryotic common ancestor".Nature Ecology & Evolution.3(3): 338–344.Bibcode:2019NatEE...3..338O.doi:10.1038/s41559-019-0796-3.hdl:10261/201794.PMID30778187.S2CID67790751.
- ^Leander BS (May 2020)."Predatory protists".Current Biology.30(10): R510–R516.Bibcode:2020CBio...30.R510L.doi:10.1016/j.cub.2020.03.052.PMID32428491.S2CID218710816.
- ^Strassert JF, Irisarri I, Williams TA, Burki F (March 2021)."A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids".Nature Communications.12(1): 1879.Bibcode:2021NatCo..12.1879S.doi:10.1038/s41467-021-22044-z.PMC7994803.PMID33767194.
- ^Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC (2013)."Molecular paleontology and complexity in the last eukaryotic common ancestor".Critical Reviews in Biochemistry and Molecular Biology.48(4): 373–396.doi:10.3109/10409238.2013.821444.PMC3791482.PMID23895660.
- ^Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo RI, et al. (2023)."Actin cytoskeleton and complex cell architecture in an Asgard archaean".Nature.613(7943): 332–339.Bibcode:2023Natur.613..332R.doi:10.1038/s41586-022-05550-y.hdl:20.500.11850/589210.PMC9834061.PMID36544020.
- ^Miao, L.; Yin, Z.; Knoll, A. H.; Qu, Y.; Zhu, M. (2024)."1.63-billion-year-old multicellular eukaryotes from the Chuanlinggou Formation in North China".Science Advances.10(4): eadk3208.Bibcode:2024SciA...10K3208M.doi:10.1126/sciadv.adk3208.PMC10807817.PMID38266082.
- ^Han TM, Runnegar B (July 1992). "Megascopic eukaryotic algae from the 2.1-billion-year-old negaunee iron-formation, Michigan".Science.257(5067): 232–5.Bibcode:1992Sci...257..232H.doi:10.1126/science.1631544.PMID1631544.
- ^Knoll AH, Javaux EJ, Hewitt D, Cohen P (June 2006)."Eukaryotic organisms in Proterozoic oceans".Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences.361(1470): 1023–1038.doi:10.1098/rstb.2006.1843.PMC1578724.PMID16754612.
- ^abcRetallack GJ, Krull ES, Thackray GD, Parkinson DH (2013). "Problematic urn-shaped fossils from a Paleoproterozoic (2.2 Ga) paleosol in South Africa".Precambrian Research.235:71–87.Bibcode:2013PreR..235...71R.doi:10.1016/j.precamres.2013.05.015.
- ^El Albani A, Bengtson S, Canfield DE, et al. (July 2010). "Large colonial organisms with coordinated growth in oxygenated environments 2.1 Gyr ago".Nature.466(7302): 100–104.Bibcode:2010Natur.466..100A.doi:10.1038/nature09166.PMID20596019.S2CID4331375.
- ^El Albani, Abderrazak (2023)."A search for life in Palaeoproterozoic marine sediments using Zn isotopes and geochemistry"(PDF).Earth and Planetary Science Letters.623:118169.Bibcode:2023E&PSL.61218169E.doi:10.1016/j.epsl.2023.118169.S2CID258360867.
- ^Ossa Ossa, Frantz; Pons, Marie-Laure; Bekker, Andrey; Hofmann, Axel; Poulton, Simon W.; et al. (2023)."Zinc enrichment and isotopic fractionation in a marine habitat of the c. 2.1 Ga Francevillian Group: A signature of zinc utilization by eukaryotes?"(PDF).Earth and Planetary Science Letters.611:118147.Bibcode:2023E&PSL.61118147O.doi:10.1016/j.epsl.2023.118147.
- ^Fakhraee, Mojtaba; Tarhan, Lidya G.; Reinhard, Christopher T.; Crowe, Sean A.; Lyons, Timothy W.; Planavsky, Noah J. (May 2023)."Earth's surface oxygenation and the rise of eukaryotic life: Relationships to the Lomagundi positive carbon isotope excursion revisited".Earth-Science Reviews.240:104398.Bibcode:2023ESRv..24004398F.doi:10.1016/j.earscirev.2023.104398.S2CID257761993.
- ^Bengtson S, Belivanova V, Rasmussen B, Whitehouse M (May 2009)."The controversial" Cambrian "fossils of the Vindhyan are real but more than a billion years older".Proceedings of the National Academy of Sciences of the United States of America.106(19): 7729–7734.Bibcode:2009PNAS..106.7729B.doi:10.1073/pnas.0812460106.PMC2683128.PMID19416859.
- ^Ward P(9 February 2008)."Mass extinctions: the microbes strike back".New Scientist.pp. 40–43.Archivedfrom the original on 8 July 2008.Retrieved27 August2017.
- ^French KL, Hallmann C, Hope JM, Schoon PL, Zumberge JA, Hoshino Y, Peters CA, George SC, Love GD, Brocks JJ, Buick R, Summons RE (May 2015)."Reappraisal of hydrocarbon biomarkers in Archean rocks".Proceedings of the National Academy of Sciences of the United States of America.112(19): 5915–5920.Bibcode:2015PNAS..112.5915F.doi:10.1073/pnas.1419563112.PMC4434754.PMID25918387.
- ^Brocks JJ, Jarrett AJ, Sirantoine E, Hallmann C, Hoshino Y, Liyanage T (August 2017). "The rise of algae in Cryogenian oceans and the emergence of animals".Nature.548(7669): 578–581.Bibcode:2017Natur.548..578B.doi:10.1038/nature23457.PMID28813409.S2CID205258987.
- ^Gold DA, Caron A, Fournier GP, Summons RE (March 2017)."Paleoproterozoic sterol biosynthesis and the rise of oxygen".Nature.543(7645): 420–423.Bibcode:2017Natur.543..420G.doi:10.1038/nature21412.hdl:1721.1/128450.PMID28264195.S2CID205254122.
- ^Wei JH, Yin X,Welander PV(24 June 2016)."Sterol Synthesis in Diverse Bacteria".Frontiers in Microbiology.7:990.doi:10.3389/fmicb.2016.00990.PMC4919349.PMID27446030.
- ^Hoshino Y, Gaucher EA (June 2021)."Evolution of bacterial steroid biosynthesis and its impact on eukaryogenesis".Proceedings of the National Academy of Sciences of the United States of America.118(25): e2101276118.Bibcode:2021PNAS..11801276H.doi:10.1073/pnas.2101276118.PMC8237579.PMID34131078.
- ^Isson TT, Love GD, Dupont CL, et al. (June 2018)."Tracking the rise of eukaryotes to ecological dominance with zinc isotopes".Geobiology.16(4): 341–352.Bibcode:2018Gbio...16..341I.doi:10.1111/gbi.12289.PMID29869832.
External links
- "Eukaryotes"Archived29 January 2012 at theWayback Machine(Tree of Life Web Project)
- "Eukaryote"at theEncyclopedia of Life