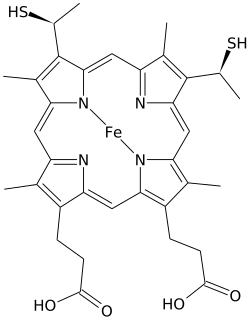
Heme C
| |
| Identifiers | |
|---|---|
3D model (JSmol)
|
|
| ChemSpider | |
| MeSH | heme+C |
PubChemCID
|
|
| |
| |
| Properties | |
| C34H36O4N4S2Fe | |
| Molar mass | 684.64904 g/mol |
Except where otherwise noted, data are given for materials in theirstandard state(at 25 °C [77 °F], 100 kPa).
| |
Heme C(orhaem C) is an important kind ofheme.
History
[edit]The correct structure of heme C was published in mid 20th century by the Swedish biochemist K.-G. Paul.[1]This work confirmed the structure first inferred by the great Swedish biochemistHugo Theorell.The structure of heme C, based upon NMR and IR experiments of the reduced Fe(II) form of the heme, was confirmed in 1975.[2]The structure of heme C including the absolutestereochemicalconfiguration about the thioether bonds was first presented for the vertebrate protein, cytochrome c[3]and is now extended to many other heme C containing proteins.
Properties
[edit]Heme C differs fromheme Bin that the twovinylside chains of heme B are replaced by covalent,thioetherlinkages to theapoprotein.The twothioetherlinkages are typically made by cysteine residues of the protein. These linkages do not allow the heme C to easily dissociate from theholoprotein,cytochrome c,compared with the more easily dissociated heme B that may dissociate from the holoprotein, the heme-protein complex, even under mild conditions. This allows a very wide range of cytochrome c structure and function, with myriad c typecytochromesacting primarily as electron carriers. The redox potential for cytochrome c can also be "fine-tuned" by small changes in protein structure and solvent interaction.[4]
The number of heme C units bound to aholoproteinis highly variable. For vertebrate cells one heme C per protein is the rule but for bacteria this number is often 2, 4, 5, 6 or even 16 heme C groups per holoprotein. It is generally agreed the number and arrangement of heme C groups are related and even required for proper holoprotein function. For instance, those proteins containing several heme C groups are involved with multiple electron transfer reactions, particularly important is the 6 electron reduction required to reduce atmospheric nitrogen into two organic ammonia molecules. It is common for the heme C to amino acid ratio to be high for bacterialhemeproteins,so the interiors of some cytochrome c proteins appear packed with many heme C groups compared with other hemeproteins. Some hemeproteins, often fromsingle cell organisms,may contain five hemes C.[5]Thebc1complexis another important enzyme that contains a C type heme.
The thioether linkages seem to allow a great freedom of function for the holoproteins. In general, the c type cytochromes can be "fine tuned" over a wider range of oxidation-reduction potential than cytochromes b. This may be an important reason why cytochrome c is nearly ubiquitous throughout life. Heme C also plays an important role inapoptosiswhere just a few molecules of cytoplasmic cytochrome c, which must still contain heme C, leads to programmed cell death.[6]Cytochrome c can be measured in human serum and can be used as a marker for inflammation.[7]
In addition to these equatorial covalent bonds, the heme iron is also usually axially coordinated to the side chains of twoamino acids,making the iron hexacoordinate. For example, mammalian and tunacytochrome ccontain a single heme C that is axially coordinated to side chains of bothhistidineandmethionine.[8]Perhaps because of the two covalent bonds holding the heme to the protein, the iron of heme C is sometimes axially ligated to the amino group oflysineor even water.
See also
[edit]References
[edit]- ^Paul, K.G.; Högfeldt, Erik; Sillén, Lars Gunnar; Kinell, Per-Olof (1950)."The splitting with silver salts of the cysteine-porphyrin bonds in cytochrome c".Acta Chemica Scandinavica.4:239–244.doi:10.3891/acta.chem.scand.04-0239.
- ^Caughey, W.S.; Smythe, G.A.; O'Keeffe, D.H.; Maskasky, J.E.; Smith, M.L. (1975)."Heme A of Cytochrome c Oxidase".Journal of Biological Chemistry.250(19): 7602–7622.doi:10.1016/S0021-9258(19)40860-0.PMID170266.
- ^Takano T.; Trus B.L.; Mandel N.; Mandel G.; Kallai O.B.; Swanson R.; Dickerson R.E. (1977)."Tuna cytochrome c at 2.0 A resolution. II. Ferrocytochrome structure analysis".Journal of Biological Chemistry.252(2): 776–785.doi:10.1016/S0021-9258(17)32784-9.PMID188826.
- ^Berghuis, A.M.; Brayer, G.D. (1992). "Oxidation state-dependent conformational changes in cytochrome c.".J. Mol. Biol.223(4): 959–976.doi:10.1016/0022-2836(92)90255-i.PMID1311391.
- ^Gwyer James D., Richardson David J., Butt Julea N. (2005). "Diode or Tunnel-Diode Characteristics? Resolving the Catalytic Consequences of Proton Coupled Electron Transfer in a Multi-Centered Oxidoreductase".Journal of the American Chemical Society.127(43): 14964–14965.doi:10.1021/ja054160s.PMID16248601.
{{cite journal}}:CS1 maint: multiple names: authors list (link) - ^Bowman, S.E.J., Bren, K.L. (2008)."The chemistry and biochemistry of heme C: functional bases for covalent attachment".Nat. Prod. Rep.25(6): 1118–1130.doi:10.1039/b717196j.PMC2654777.PMID19030605.
{{cite journal}}:CS1 maint: multiple names: authors list (link) - ^Eleftheriadis, T.; Pissas, G.; Liakopoulos, V.; Stafanidis, I. (2016)."Cytochrome c as a Potentially Clinical Useful Marker of Mitochondrial and Cellular Damage".Front. Immunol.7:279.doi:10.3389/fimmu.2016.00279.PMC4951490.PMID27489552.
- ^Yeh, S.R., Han, S., and Rousseau, D.L. (1998). "Cytochrome c folding and unfolding".Accounts of Chemical Research.31(11): 727–735.doi:10.1021/ar970084p.
{{cite journal}}:CS1 maint: multiple names: authors list (link)
