Heterochromatin
Heterochromatinis a tightly packed form ofDNAorcondensed DNA,which comes in multiple varieties. These varieties lie on a continuum between the two extremes ofconstitutive heterochromatinandfacultative heterochromatin.Both play a role in theexpression of genes.Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002),[1]and many other papers since,[2]much of this DNA is in fact transcribed, but it is continuouslyturned overviaRNA-induced transcriptional silencing(RITS). Recent studies withelectron microscopyandOsO4staining reveal that the dense packing is not due to the chromatin.[3]
Constitutive heterochromatin can affect the genes near itself (e.g.position-effect variegation). It is usuallyrepetitiveand forms structural functions such ascentromeresortelomeres,in addition to acting as an attractor for other gene-expression or repression signals.
Facultative heterochromatin is the result of genes that aresilencedthrough a mechanism such ashistone deacetylationorPiwi-interacting RNA(piRNA) throughRNAi.It is not repetitive and shares the compact structure of constitutive heterochromatin. However, under specific developmental or environmental signaling cues, it can lose its condensed structure and become transcriptionally active.[4]
Heterochromatin has been associated with thedi-andtri-methylation ofH3K9in certain portions of the human genome.[5]H3K9me3-relatedmethyltransferasesappear to have a pivotal role in modifying heterochromatin during lineage commitment at the onset oforganogenesisand in maintaining lineage fidelity.[6]
Structure
[edit]
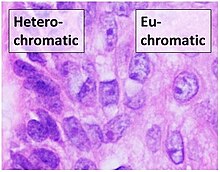
Chromatinis found in two varieties:euchromatinand heterochromatin.[7]Originally, the two forms were distinguished cytologically by how intensely they get stained – the euchromatin is less intense, while heterochromatin stains intensely, indicating tighter packing. Heterochromatin was given its name for this reason by botanist Emil Heitz who discovered that heterochromatin remained darkly stained throughout the entire cell cycle, unlike euchromatin whose stain disappeared during interphase.[8]Heterochromatin is usually localized to the periphery of thenucleus. Despite this early dichotomy, recent evidence in both animals[9]and plants[10]has suggested that there are more than two distinct heterochromatin states, and it may in fact exist in four or five 'states', each marked by different combinations ofepigeneticmarks.
Heterochromatin mainly consists of genetically inactivesatellite sequences,[11]and many genes are repressed to various extents, although some cannot be expressed in euchromatin at all.[12]Bothcentromeresandtelomeresare heterochromatic, as is theBarr bodyof the second, inactivatedX-chromosomein a female.
Function
[edit]
Heterochromatin has been associated with several functions, from gene regulation to the protection of chromosome integrity;[13]some of these roles can be attributed to the dense packing of DNA, which makes it less accessible to protein factors that usually bind DNA or its associated factors. For example, naked double-stranded DNA ends would usually be interpreted by the cell as damaged or viral DNA, triggeringcell cyclearrest,DNA repairor destruction of the fragment, such as byendonucleasesin bacteria.
Some regions of chromatin are very densely packed with fibers that display a condition comparable to that of the chromosome atmitosis.Heterochromatin is generally clonally inherited; when a cell divides, the two daughter cells typically contain heterochromatin within the same regions of DNA, resulting inepigenetic inheritance.Variations cause heterochromatin to encroach on adjacent genes or recede from genes at the extremes of domains. Transcribable material may be repressed by being positioned (incis) at these boundary domains. This gives rise to expression levels that vary from cell to cell,[14]which may be demonstrated byposition-effect variegation.[15]Insulatorsequences may act as a barrier in rare cases where constitutive heterochromatin and highly active genes are juxtaposed (e.g. the 5'HS4 insulator upstream of the chicken β-globin locus,[16]and loci in twoSaccharomycesspp.[17][18]).
Constitutive heterochromatin
[edit]All cells of a given species package the same regions of DNA inconstitutive heterochromatin,and thus in all cells, any genes contained within the constitutive heterochromatin will be poorlyexpressed.For example, all human chromosomes1,9,16,and theY-chromosomecontain large regions of constitutive heterochromatin. In most organisms, constitutive heterochromatin occurs around the chromosome centromere and near telomeres.
Facultative heterochromatin
[edit]
The regions of DNA packaged in facultative heterochromatin will not be consistent between the cell types within a species, and thus a sequence in one cell that is packaged in facultative heterochromatin (and the genes within are poorly expressed) may be packaged in euchromatin in another cell (and the genes within are no longer silenced). However, the formation of facultative heterochromatin is regulated, and is often associated withmorphogenesisordifferentiation.An example of facultative heterochromatin isX chromosome inactivationin female mammals: oneX chromosomeis packaged as facultative heterochromatin and silenced, while the other X chromosome is packaged as euchromatin and expressed.
Among the molecular components that appear to regulate the spreading of heterochromatin are thePolycomb-group proteinsand non-coding genes such asXist.The mechanism for such spreading is still a matter of controversy.[19]The polycomb repressive complexesPRC1andPRC2regulatechromatincompaction and gene expression and have a fundamental role in developmental processes. PRC-mediatedepigeneticaberrations are linked togenome instabilityand malignancy and play a role in theDNA damageresponse,DNA repairand in the fidelity ofreplication.[20]
Yeast heterochromatin
[edit]Saccharomyces cerevisiae,or budding yeast, is a modeleukaryoteand its heterochromatin has been defined thoroughly. Although most of its genome can be characterized as euchromatin,S. cerevisiaehas regions of DNA that are transcribed very poorly. These loci are the so-called silent mating type loci (HML and HMR), the rDNA (encoding ribosomal RNA), and the sub-telomeric regions. Fission yeast (Schizosaccharomyces pombe) uses another mechanism for heterochromatin formation at its centromeres. Gene silencing at this location depends on components of theRNAipathway. Double-stranded RNA is believed to result in silencing of the region through a series of steps.
In the fission yeastSchizosaccharomyces pombe,two RNAi complexes, the RITS complex and the RNA-directed RNA polymerase complex (RDRC), are part of an RNAi machinery involved in the initiation, propagation and maintenance of heterochromatin assembly. These two complexes localize in asiRNA-dependent manner on chromosomes, at the site of heterochromatin assembly.RNA polymerase IIsynthesizes a transcript that serves as a platform to recruit RITS, RDRC and possibly other complexes required for heterochromatin assembly.[21][22]Both RNAi and an exosome-dependent RNA degradation process contribute to heterochromatic gene silencing. These mechanisms ofSchizosaccharomyces pombemay occur in other eukaryotes.[23]A large RNA structure calledRevCenhas also been implicated in the production of siRNAs to mediate heterochromatin formation in some fission yeast.[24]
See also
[edit]References
[edit]- ^Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (September 2002)."Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi".Science.297(5588): 1833–7.Bibcode:2002Sci...297.1833V.doi:10.1126/science.1074973.PMID12193640.S2CID2613813.
- ^"What is the current evidence showing active transcription withinin..."researchgate.net.Retrieved2016-04-30.
- ^Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC (July 2017)."ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells".Science.357(6349): eaag0025.doi:10.1126/science.aag0025.PMC5646685.PMID28751582.
- ^Oberdoerffer P, Sinclair DA (September 2007)."The role of nuclear architecture in genomic instability and ageing".Nature Reviews. Molecular Cell Biology.8(9): 692–702.doi:10.1038/nrm2238.PMID17700626.S2CID15674132.
- ^Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ (March 2009)."Determination of enriched histone modifications in non-genic portions of the human genome".BMC Genomics.10(1): 143.doi:10.1186/1471-2164-10-143.PMC2667539.PMID19335899.
- ^Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, et al. (January 2019)."H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification".Science.363(6424): 294–297.Bibcode:2019Sci...363..294N.doi:10.1126/science.aau0583.PMC6664818.PMID30606806.
- ^ Elgin, S.C. (1996)."Heterochromatin and gene regulation inDrosophila".Current Opinion in Genetics & Development.6(2): 193–202.doi:10.1016/S0959-437X(96)80050-5.ISSN0959-437X.PMID8722176.
- ^Penagos-Puig, Andrés; Furlan-Magaril, Mayra (2020-09-18)."Heterochromatin as an Important Driver of Genome Organization".Frontiers in Cell and Developmental Biology.8.doi:10.3389/fcell.2020.579137.ISSN2296-634X.PMC7530337.PMID33072761.
- ^ van Steensel B (May 2011)."Chromatin: constructing the big picture".The EMBO Journal.30(10): 1885–95.doi:10.1038/emboj.2011.135.PMC3098493.PMID21527910.
- ^ Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, et al. (May 2011)."Integrative epigenomic mapping defines four main chromatin states in Arabidopsis".The EMBO Journal.30(10): 1928–38.doi:10.1038/emboj.2011.103.PMC3098477.PMID21487388.
- ^ Lohe AR, Hilliker AJ, Roberts PA (August 1993)."Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster".Genetics.134(4): 1149–74.doi:10.1093/genetics/134.4.1149.PMC1205583.PMID8375654.
- ^ Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC (June 2000)."Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila".Genetics.155(2): 699–708.doi:10.1093/genetics/155.2.699.PMC1461102.PMID10835392.
- ^
Grewal SI, Jia S (January 2007)."Heterochromatin revisited".Nature Reviews. Genetics.8(1): 35–46.doi:10.1038/nrg2008.PMID17173056.S2CID31811880.
An up-to-date account of the current understanding of repetitive DNA, which usually doesn't contain genetic information. If evolution makes sense only in the context of the regulatory control of genes, we propose that heterochromatin, which is the main form of chromatin in higher eukaryotes, is positioned to be a deeply effective target for evolutionary change. Future investigations into assembly, maintenance and the many other functions of heterochromatin will shed light on the processes of gene and chromosome regulation.
- ^ Fisher AG, Merkenschlager M (April 2002). "Gene silencing, cell fate and nuclear organisation".Current Opinion in Genetics & Development.12(2): 193–7.doi:10.1016/S0959-437X(02)00286-1.PMID11893493.
- ^ Zhimulev, I.F.[in Russian];et al. (December 1986). "Cytogenetic and molecular aspects of position effect variegation in Drosophila melanogaster".Chromosoma.94(6): 492–504.doi:10.1007/BF00292759.ISSN1432-0886.S2CID24439936.
- ^ Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, et al. (December 2002)."The insulation of genes from external enhancers and silencing chromatin".Proceedings of the National Academy of Sciences of the United States of America.99(Suppl 4): 16433–7.Bibcode:2002PNAS...9916433B.doi:10.1073/pnas.162342499.PMC139905.PMID12154228.
- ^ Allis CD, Grewal SI (August 2001). "Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries".Science.293(5532): 1150–5.doi:10.1126/science.1064150.PMID11498594.S2CID26350729.
- ^ Donze D, Kamakaka RT (February 2001)."RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae".The EMBO Journal.20(3): 520–31.doi:10.1093/emboj/20.3.520.PMC133458.PMID11157758.
- ^Talbert PB, Henikoff S (October 2006). "Spreading of silent chromatin: inaction at a distance".Nature Reviews. Genetics.7(10): 793–803.doi:10.1038/nrg1920.PMID16983375.S2CID1671107.
- ^Veneti Z, Gkouskou KK, Eliopoulos AG (July 2017)."Polycomb Repressor Complex 2 in Genomic Instability and Cancer".International Journal of Molecular Sciences.18(8): 1657.doi:10.3390/ijms18081657.PMC5578047.PMID28758948.
- ^Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (July 2005). "RNA polymerase II is required for RNAi-dependent heterochromatin assembly".Science.309(5733): 467–9.Bibcode:2005Sci...309..467K.doi:10.1126/science.1114955.PMID15947136.S2CID22636283.
- ^Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (October 2005)."RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing".Genes & Development.19(19): 2301–6.doi:10.1101/gad.344205.PMC1240039.PMID16204182.
- ^Vavasseur; et al. (2008)."Heterochromatin Assembly and Transcriptional Gene Silencing under the Control of Nuclear RNAi: Lessons from Fission Yeast".RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity.Caister Academic Press.ISBN978-1-904455-25-7.
- ^Djupedal I, Kos-Braun IC, Mosher RA, Söderholm N, Simmer F, Hardcastle TJ, et al. (December 2009)."Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA".The EMBO Journal.28(24): 3832–44.doi:10.1038/emboj.2009.351.PMC2797062.PMID19942857.
External links
[edit]- Histology image: 20102loa– Histology Learning System at Boston University
- Avramova ZV (May 2002)."Heterochromatin in animals and plants. Similarities and differences".Plant Physiology.129(1): 40–9.doi:10.1104/pp.010981.PMC1540225.PMID12011336.
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, et al. (February 2001)."The human transcriptome map: clustering of highly expressed genes in chromosomal domains".Science.291(5507): 1289–92.Bibcode:2001Sci...291.1289C.doi:10.1126/science.1056794.PMID11181992.
- Cha, Ariana Eunjung; Bernstein, Lenny (April 30, 2015)."Scientists discover an important new driver of aging".New York Times.Retrieved4 May2015.
