Phycosphere
| Part of a series on |
| Microbiomes |
|---|
 |
Thephycosphereis a microscale mucus region that is rich inorganic mattersurrounding aphytoplanktoncell. This area is high in nutrients due to extracellular waste from the phytoplankton cell and it has been suggested thatbacteriainhabit this area to feed on these nutrients. This high nutrient environment creates amicrobiomeand a diversefood webfor microbes such asbacteriaandprotists.[1]It has also been suggested that the bacterial assemblages within the phycosphere are species-specific and can vary depending on differentenvironmental factors.[2]
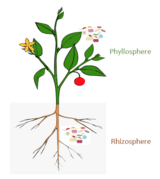
In terms of comparison, the phycosphere inphytoplanktonhas been suggested analogous to therhizosphereinplants,which is the root zone important for nutrient recycling. Both plant roots and phytoplankton exude chemicals which alter their immediate surrounds drastically – including altering the pH and oxygen levels. In terms of community construction, chemotaxis is used in both environments in order to propagate the recruitment of microbes. In the rhizosphere, chemotaxis is used by the host – the plant – to mediate the motility of the soil which allows for microbial colonization. In the phycosphere, the phytoplankton release of specific chemical exudates elicits a response from bacterial symbionts who exhibit chemotaxis signaling, thereby enabling the recruitment of microbes and subsequent colonization. The interfaces also have a few similar microbes, chemicals, and metabolites involved in the host – symbiont interactions. This includes microbes such as Rhizobium, which in the phycospheres of green algae was found to be the foremost microbe when compared to other abundant community members. Chemicals such as dimethylsuloniopropionate (DMSP) and 2,3-dihydroxypropane-1-sulfonate (DHPS) and metabolites such as sugars and amino acids are implicated in the mechanisms of action of both microbiomes.
Phytoplankton-bacteria interactions
[edit]Themicroscaleinteractions between thephytoplanktonandbacteriaare complex. The phytoplankton-bacteria interactions have the potential to beparasitism,competition ormutualism.
Interactions between phytoplankton and bacteria in the phycosphere could be potentially important in low-nutrient regions of the ocean and an example of mutualism. In marineecosystemsthat are low in nutrients (i.e.oligotrophicregions of the oceans), it could be potentially beneficial for the phytoplankton to have remineralizingbacteriain the phycosphere fornutrientrecycling.It has been suggested that while the bacterial activity may be low, thetaxonomicdiversityand nutritionaldiversityis high.[3]This can possibly suggest that the phytoplankton species may rely on a diverse array of bacterial interactions for recyclednutrientsin theseoligotrophicregions and the bacteria rely on organic matter surrounding the phycosphere for a source of food.
However, bacterial-phytoplankton interactions in the phycosphere could beparasitic.In the same low nutrient oligotrophic regions of the ocean,phytoplanktonthat arenutrientstressed may not be able to produce this protective mucus layer or its associatedantibiotics.Thebacteria,who are also food stressed, could kill thephytoplanktonand use it as a food substrate.[4]
Also,bacteriametabolize theorganic matterthroughaerobic respiration,which depletes oxygen from the water and can lower thepHof the water column. If enoughorganic matteris produced, the bacteria could potentially harm the phytoplankton by causing the water to become moreacidic.(See alsoeutrophication).
Examples of bacteria associated with phycosphere
[edit]In reality, the actualbacterialdiversityof the phycosphere is extremely diverse and is dependent environmental factors, such asturbulencein the water (so the bacteria can attach to the mucus or the phytoplankton cell) or the concentrations of nutrients. Also, the bacteria tend to be highly specialized when associated with this region. Nevertheless, here are some examples ofbacteriumgeneraassociated with the phycosphere.
- Pseudomonas
- Achromobacter
- Roseobacter
- Flavobacteraceae
- Alteromonadaceae
- Athrospira plantensis
- Terrimonas rubra
- C.vulgaris
- Sediminibacterium
- Chryseobacterium
See also
[edit]References
[edit]- ^Sigee, David (2005).Freshwater Microbiology: Biodiversity and Dynamic Interactions of Microorganisms in the Aquatic Environment.West Sussex, England: John Wiley & Sons.ISBN978-0-471-48529-2.
- ^Sapp, Melanie; Schwaderer, Anne S.; Wiltshire, Karen H.; Hoppe, Hans-Georg; Gerdts, Gunnar; Wichels, Antje (31 January 2007). "Species-Specific Bacterial Communities in the Phycosphere of Microalgae?".Microbial Ecology.53(4): 683–699.doi:10.1007/s00248-006-9162-5.PMID17264999.S2CID21278078.
- ^Cole, Jonathan J. (1982). "Interactions Between Bacteria and Algae in Aquatic Ecosystems".Annual Review of Ecology and Systematics.13:291–314.doi:10.1146/annurev.es.13.110182.001451.
- ^Azam, Farooq; Malfatti, Francesca (October 2007). "Microbial structuring of marine ecosystems".Nature.5(10): 782–791.doi:10.1038/nrmicro1747.PMID17853906.S2CID10055219.
5. Seymour, Justin R., et al. “Zooming in on the Phycosphere: the Ecological Interface for Phytoplankton–Bacteria Relationships.” Nature Microbiology, vol. 2, no. 7, 2017, pp. 1–13., doi:10.1038/nmicrobiol.2017.65.
6. Kim, B.-H., Ramanan, R., Cho, D.-H., Oh, H.-M., & Kim, H.-S. (2014). Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass and Bioenergy, 69, 95–105. doi: 10.1016/j.biombioe.2014.07.015
7. Geng, H., & Belas, R. (2010). Molecular mechanisms underlying roseobacter–phytoplankton symbioses. Current Opinion in Biotechnology, 21(3), 332–338. doi: 10.1016/j.copbio.2010.03.013 8. Ramanan, R., Kang, Z., Kim, B.-H., Cho, D.-H., Jin, L., Oh, H.-M., & Kim, H.-S. (2015). Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Research, 8, 140–144. doi: 10.1016/j.algal.2015.02.003
9. Scharf, B. E., Hynes, M. F., & Alexandre, G. M. (2016). Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant Molecular Biology, 90(6), 549–559. doi: 10.1007/s11103-016-0432-4
