Polyadenylation

Polyadenylationis the addition of apoly(A) tailto an RNA transcript, typically amessenger RNA(mRNA). The poly(A) tail consists of multipleadenosine monophosphates;in other words, it is a stretch of RNA that has onlyadeninebases. Ineukaryotes,polyadenylation is part of the process that produces mature mRNA fortranslation.In manybacteria,the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process ofgene expression.
The process of polyadenylation begins as thetranscriptionof ageneterminates.The3′-mostsegment of the newly made pre-mRNA is first cleaved off by aset of proteins;these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar toalternative splicing.[1]
The poly(A) tail is important for the nuclear export, translation and stability of mRNA. The tail is shortened over time, and, when it is short enough, the mRNA is enzymatically degraded.[2]However, in a few cell types, mRNAs with short poly(A) tails are stored for later activation by re-polyadenylation in the cytosol.[3]In contrast, when polyadenylation occurs in bacteria, it promotes RNA degradation.[4]This is also sometimes the case for eukaryoticnon-coding RNAs.[5][6]
mRNA molecules in both prokaryotes and eukaryotes have polyadenylated 3′-ends, with the prokaryotic poly(A) tails generally shorter and fewer mRNA molecules polyadenylated.[7]
Background on RNA
[edit]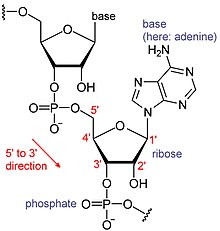
RNAs are a type of large biological molecules, whose individual building blocks are called nucleotides. The namepoly(A) tail(for polyadenylic acid tail)[8]reflects the way RNA nucleotides are abbreviated, with a letter for the base the nucleotide contains (A foradenine,C forcytosine,G forguanineand U foruracil). RNAs are produced (transcribed) from aDNAtemplate. By convention, RNA sequences are written in a 5′ to 3′ direction. The 5′ end is the part of the RNA molecule that is transcribed first, and the 3′ end is transcribed last. The 3′ end is also where the poly(A) tail is found on polyadenylated RNAs.[1][9]
Messenger RNA (mRNA) is RNA that has a coding region that acts as a template for protein synthesis (translation). The rest of the mRNA, theuntranslated regions,tune how active the mRNA is.[10]There are also many RNAs that are not translated, called non-coding RNAs. Like the untranslated regions, many of these non-coding RNAs have regulatory roles.[11]
Nuclear polyadenylation
[edit]Function
[edit]In nuclear polyadenylation, a poly(A) tail is added to an RNA at the end of transcription. On mRNAs, the poly(A) tail protects the mRNA molecule from enzymatic degradation in thecytoplasmand aids in transcription termination, export of the mRNA from the nucleus, and translation.[2]Almost all eukaryotic mRNAs are polyadenylated,[12]with the exception of animal replication-dependenthistonemRNAs.[13]These are the only mRNAs in eukaryotes that lack a poly(A) tail, ending instead in astem-loopstructure followed by a purine-rich sequence, termed histone downstream element, that directs where the RNA is cut so that the 3′ end of the histone mRNA is formed.[14]
Many eukaryotic non-coding RNAs are always polyadenylated at the end of transcription. There are small RNAs where the poly(A) tail is seen only in intermediary forms and not in the mature RNA as the ends are removed during processing, the notable ones beingmicroRNAs.[15][16]But, for manylong noncoding RNAs– a seemingly large group ofregulatoryRNAs that, for example, includes the RNAXist,which mediatesX chromosome inactivation– a poly(A) tail is part of the mature RNA.[17]
Mechanism
[edit]| Proteins involved:[12][18] CPSF:cleavage/polyadenylation specificity factor |
Theprocessivepolyadenylation complex in the nucleus of eukaryotes works on products ofRNA polymerase II,such asprecursor mRNA.Here, a multi-protein complex(see components on the right)[18]cleaves the 3′-most part of a newly produced RNA and polyadenylates the end produced by this cleavage. The cleavage is catalysed by the enzymeCPSF[13][18]and occurs 10–30 nucleotides downstream of its binding site.[19]This site often has thepolyadenylation signalsequence AAUAAA on the RNA, but variants of it that bind more weakly toCPSFexist.[18][20]Two other proteins add specificity to the binding to an RNA: CstF and CFI. CstF binds to a GU-rich region further downstream of CPSF's site.[21]CFI recognises a third site on the RNA (a set of UGUAA sequences in mammals[22][23][24]) and can recruit CPSF even if the AAUAAA sequence is missing.[25][26]The polyadenylation signal – the sequence motif recognised by the RNA cleavage complex – varies between groups of eukaryotes. Most human polyadenylation sites contain the AAUAAA sequence,[21]but this sequence is less common in plants and fungi.[27]
The RNA is typically cleaved before transcription termination, as CstF also binds to RNA polymerase II.[28]Through a poorly understood mechanism (as of 2002), it signals for RNA polymerase II to slip off of the transcript.[29]Cleavage also involves the protein CFII, though it is unknown how.[30]The cleavage site associated with a polyadenylation signal can vary up to some 50 nucleotides.[31]
When the RNA is cleaved, polyadenylation starts, catalysed by polyadenylate polymerase.Polyadenylate polymerasebuilds the poly(A) tail by addingadenosine monophosphateunits fromadenosine triphosphateto the RNA, cleaving offpyrophosphate.[32]Another protein, PAB2, binds to the new, short poly(A) tail and increases the affinity of polyadenylate polymerase for the RNA. When the poly(A) tail is approximately 250nucleotideslong the enzyme can no longer bind to CPSF and polyadenylation stops, thus determining the length of the poly(A) tail.[33][34]CPSF is in contact with RNA polymerase II, allowing it to signal the polymerase to terminate transcription.[35][36]When RNA polymerase II reaches a "termination sequence" (⁵'TTTATT3' on the DNA template and ⁵'AAUAAA3' on the primary transcript), the end of transcription is signaled.[37]The polyadenylation machinery is also physically linked to thespliceosome,a complex that removesintronsfrom RNAs.[26]
Downstream effects
[edit]The poly(A) tail acts as the binding site forpoly(A)-binding protein.Poly(A)-binding protein promotes export from the nucleus and translation, and inhibits degradation.[38]This protein binds to the poly(A) tail prior to mRNA export from the nucleus and in yeast also recruits poly(A) nuclease, an enzyme that shortens the poly(A) tail and allows the export of the mRNA. Poly(A)-binding protein is exported to the cytoplasm with the RNA. mRNAs that are not exported are degraded by theexosome.[39][40]Poly(A)-binding protein also can bind to, and thus recruit, several proteins that affect translation,[39]one of these isinitiation factor-4G, which in turn recruits the40Sribosomal subunit.[41]However, a poly(A) tail is not required for the translation of all mRNAs.[42]Further, poly(A) tailing (oligo-adenylation) can determine the fate of RNA molecules that are usually not poly(A)-tailed (such as (small) non-coding (sn)RNAs etc.) and thereby induce their RNA decay.[43]
Deadenylation
[edit]In eukaryoticsomatic cells,the poly(A) tails of most mRNAs in the cytoplasm gradually get shorter, and mRNAs with shorter poly(A) tail are translated less and degraded sooner.[44]However, it can take many hours before an mRNA is degraded.[45]This deadenylation and degradation process can be accelerated by microRNAs complementary to the3′ untranslated regionof an mRNA.[46]Inimmature egg cells,mRNAs with shortened poly(A) tails are not degraded, but are instead stored and translationally inactive. These short tailed mRNAs are activated by cytoplasmic polyadenylation after fertilisation, duringegg activation.[47]
In animals, poly(A) ribonuclease (PARN) can bind to the5′ capand remove nucleotides from the poly(A) tail. The level of access to the 5′ cap and poly(A) tail is important in controlling how soon the mRNA is degraded. PARN deadenylates less if the RNA is bound by the initiation factors 4E (at the 5′ cap) and 4G (at the poly(A) tail), which is why translation reduces deadenylation. The rate of deadenylation may also be regulated by RNA-binding proteins. Additionally, RNA triple helix structures and RNA motifs such as the poly(A) tail 3’ end binding pocket retard deadenylation process and inhibit poly(A) tail removal.[48]Once the poly(A) tail is removed, the decapping complex removes the 5′ cap, leading to a degradation of the RNA. Several other proteins are involved in deadenylation inbudding yeastand human cells, most notably theCCR4-Notcomplex.[49]
Cytoplasmic polyadenylation
[edit]There is polyadenylation in the cytosol of some animal cell types, namely in thegermline,during earlyembryogenesisand in post-synapticsites ofnerve cells.This lengthens the poly(A) tail of an mRNA with a shortened poly(A) tail, so that the mRNA will betranslated.[44][50]These shortened poly(A) tails are often less than 20 nucleotides, and are lengthened to around 80–150 nucleotides.[3]
In the early mouse embryo, cytoplasmic polyadenylation of maternal RNAs from the egg cell allows the cell to survive and grow even though transcription does not start until the middle of the 2-cell stage (4-cell stage in human).[51][52]In the brain, cytoplasmic polyadenylation is active during learning and could play a role inlong-term potentiation,which is the strengthening of the signal transmission from a nerve cell to another in response to nerve impulses and is important for learning and memory formation.[3][53]
Cytoplasmic polyadenylation requires the RNA-binding proteinsCPSFandCPEB,and can involve other RNA-binding proteins likePumilio.[54]Depending on the cell type, the polymerase can be the same type of polyadenylate polymerase (PAP) that is used in the nuclear process, or the cytoplasmic polymeraseGLD-2.[55]
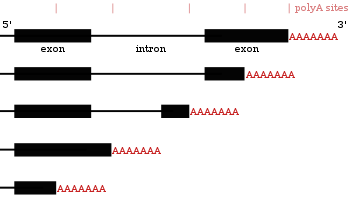
Alternative polyadenylation
[edit]Many protein-coding genes have more than one polyadenylation site, so a gene can code for several mRNAs that differ in their3′ end.[27][56][57]The 3’ region of a transcript contains many polyadenylation signals (PAS). When more proximal (closer towards 5’ end) PAS sites are utilized, this shortens the length of the 3’ untranslated region (3' UTR) of a transcript.[58]Studies in both humans and flies have shown tissue specific APA. With neuronal tissues preferring distal PAS usage, leading to longer 3’ UTRs and testis tissues preferring proximal PAS leading to shorter 3’ UTRs.[59][60]Studies have shown there is a correlation between a gene's conservation level and its tendency to do alternative polyadenylation, with highly conserved genes exhibiting more APA. Similarly, highly expressed genes follow this same pattern.[61]Ribo-sequencingdata (sequencing of only mRNAs inside ribosomes) has shown that mRNA isoforms with shorter 3’ UTRs are more likely to be translated.[58]
Since alternative polyadenylation changes the length of the3' UTR,[62]it can also change which binding sites are available formicroRNAsin the 3′ UTR.[19][63]MicroRNAs tend to repress translation and promote degradation of the mRNAs they bind to, although there are examples of microRNAs that stabilise transcripts.[64][65]Alternative polyadenylation can also shorten the coding region, thus making the mRNA code for a different protein,[66][67]but this is much less common than just shortening the 3′ untranslated region.[27]
The choice of poly(A) site can be influenced by extracellular stimuli and depends on the expression of the proteins that take part in polyadenylation.[68][69]For example, the expression ofCstF-64,a subunit ofcleavage stimulatory factor(CstF), increases inmacrophagesin response tolipopolysaccharides(a group of bacterial compounds that trigger an immune response). This results in the selection of weak poly(A) sites and thus shorter transcripts. This removes regulatory elements in the 3′ untranslated regions of mRNAs for defense-related products likelysozymeandTNF-α.These mRNAs then have longer half-lives and produce more of these proteins.[68]RNA-binding proteins other than those in the polyadenylation machinery can also affect whether a polyadenylation site is used,[70][71][72][73]as canDNA methylationnear the polyadenylation signal.[74]In addition, numerous other components involved in transcription, splicing or other mechanisms regulating RNA biology can affect APA.[75]
Tagging for degradation in eukaryotes
[edit]For manynon-coding RNAs,includingtRNA,rRNA,snRNA,andsnoRNA,polyadenylation is a way of marking the RNA for degradation, at least inyeast.[76]This polyadenylation is done in the nucleus by theTRAMP complex,which maintains a tail that is around 4 nucleotides long to the 3′ end.[77][78]The RNA is then degraded by theexosome.[79]Poly(A) tails have also been found on human rRNA fragments, both the form of homopolymeric (A only) and heterpolymeric (mostly A) tails.[80]
In prokaryotes and organelles
[edit]
In many bacteria, both mRNAs and non-coding RNAs can be polyadenylated. This poly(A) tail promotes degradation by thedegradosome,which contains two RNA-degrading enzymes:polynucleotide phosphorylaseandRNase E.Polynucleotide phosphorylase binds to the 3′ end of RNAs and the 3′ extension provided by the poly(A) tail allows it to bind to the RNAs whosesecondary structurewould otherwise block the 3′ end. Successive rounds of polyadenylation and degradation of the 3′ end by polynucleotide phosphorylase allows thedegradosometo overcome these secondary structures. The poly(A) tail can also recruit RNases that cut the RNA in two.[81]These bacterial poly(A) tails are about 30 nucleotides long.[82]
In as different groups as animals andtrypanosomes,themitochondriacontain both stabilising and destabilising poly(A) tails. Destabilising polyadenylation targets both mRNA and noncoding RNAs. The poly(A) tails are 43 nucleotides long on average. The stabilising ones start at the stop codon, and without them the stop codon (UAA) is not complete as the genome only encodes the U or UA part. Plant mitochondria have only destabilising polyadenylation. Mitochondrial polyadenylation has never been observed in either budding or fission yeast.[83][84]
While many bacteria and mitochondria have polyadenylate polymerases, they also have another type of polyadenylation, performed bypolynucleotide phosphorylaseitself. This enzyme is found in bacteria,[85]mitochondria,[86]plastids[87]and as a constituent of the archaeal exosome (in thosearchaeathat have anexosome).[88]It can synthesise a 3′ extension where the vast majority of the bases are adenines. Like in bacteria, polyadenylation by polynucleotide phosphorylase promotes degradation of the RNA in plastids[89]and likely also archaea.[83]
Evolution
[edit]Although polyadenylation is seen in almost all organisms, it is not universal.[7][90]However, the wide distribution of this modification and the fact that it is present in organisms from all threedomainsof life implies that thelast universal common ancestorof all living organisms, it is presumed, had some form of polyadenylation system.[82]A few organisms do not polyadenylate mRNA, which implies that they have lost their polyadenylation machineries during evolution. Although no examples of eukaryotes that lack polyadenylation are known, mRNAs from the bacteriumMycoplasma gallisepticumand the salt-tolerant archaeanHaloferax volcaniilack this modification.[91][92]
The most ancient polyadenylating enzyme ispolynucleotide phosphorylase.This enzyme is part of both the bacterialdegradosomeand the archaealexosome,[93]two closely related complexes that recycle RNA into nucleotides. This enzyme degrades RNA by attacking the bond between the 3′-most nucleotides with a phosphate, breaking off a diphosphate nucleotide. This reaction is reversible, and so the enzyme can also extend RNA with more nucleotides. The heteropolymeric tail added by polynucleotide phosphorylase is very rich in adenine. The choice of adenine is most likely the result of higherADPconcentrations than other nucleotides as a result of usingATPas an energy currency, making it more likely to be incorporated in this tail in early lifeforms. It has been suggested that the involvement of adenine-rich tails in RNA degradation prompted the later evolution of polyadenylate polymerases (the enzymes that produce poly(A) tails with no other nucleotides in them).[94]
Polyadenylate polymerases are not as ancient. They have separately evolved in both bacteria and eukaryotes fromCCA-adding enzyme,which is the enzyme that completes the 3′ ends oftRNAs.Its catalytic domain is homologous to that of otherpolymerases.[79]It is presumed that the horizontal transfer of bacterial CCA-adding enzyme to eukaryotes allowed the archaeal-like CCA-adding enzyme to switch function to a poly(A) polymerase.[82]Some lineages, likearchaeaandcyanobacteria,never evolved a polyadenylate polymerase.[94]
Polyadenylate tails are observed in severalRNA viruses,includingInfluenza A,[95]Coronavirus,[96]Alfalfa mosaic virus,[97]andDuck Hepatitis A.[98]Some viruses, such asHIV-1andPoliovirus,inhibit the cell's poly-A binding protein (PABPC1) in order to emphasize their own genes' expression over the host cell's.[99]
History
[edit]Poly(A)polymerase was first identified in 1960 as anenzymatic activityin extracts made from cell nuclei that could polymerise ATP, but not ADP, into polyadenine.[100][101]Although identified in many types of cells, this activity had no known function until 1971, when poly(A) sequences were found in mRNAs.[102][103]The only function of these sequences was thought at first to be protection of the 3′ end of the RNA from nucleases, but later the specific roles of polyadenylation in nuclear export and translation were identified. The polymerases responsible for polyadenylation were first purified and characterized in the 1960s and 1970s, but the large number of accessory proteins that control this process were discovered only in the early 1990s.[102]
See also
[edit]References
[edit]- ^abProudfoot NJ, Furger A, Dye MJ (February 2002)."Integrating mRNA processing with transcription".Cell.108(4): 501–12.doi:10.1016/S0092-8674(02)00617-7.PMID11909521.S2CID478260.
- ^abGuhaniyogi J, Brewer G (March 2001)."Regulation of mRNA stability in mammalian cells".Gene.265(1–2): 11–23.doi:10.1016/S0378-1119(01)00350-X.PMC3340483.PMID11255003.
- ^abcRichter JD (June 1999)."Cytoplasmic polyadenylation in development and beyond".Microbiology and Molecular Biology Reviews.63(2): 446–56.doi:10.1128/MMBR.63.2.446-456.1999.PMC98972.PMID10357857.
- ^Steege DA (August 2000)."Emerging features of mRNA decay in bacteria".RNA.6(8): 1079–90.doi:10.1017/S1355838200001023.PMC1369983.PMID10943888.
- ^Zhuang Y, Zhang H, Lin S (June 2013). "Polyadenylation of 18S rRNA in algae(1)".Journal of Phycology.49(3): 570–9.Bibcode:2013JPcgy..49..570Z.doi:10.1111/jpy.12068.PMID27007045.S2CID19863143.
- ^Anderson JT (August 2005)."RNA turnover: unexpected consequences of being tailed".Current Biology.15(16): R635-8.Bibcode:2005CBio...15.R635A.doi:10.1016/j.cub.2005.08.002.PMID16111937.S2CID19003617.
- ^abSarkar N (June 1997). "Polyadenylation of mRNA in prokaryotes".Annual Review of Biochemistry.66(1): 173–97.doi:10.1146/annurev.biochem.66.1.173.PMID9242905.
- ^Stevens A (1963). "Ribonucleic Acids-Biosynthesis and Degradation".Annual Review of Biochemistry.32:15–42.doi:10.1146/annurev.bi.32.070163.000311.PMID14140701.
- ^Lehninger AL, Nelson DL, Cox MM, eds. (1993).Principles of biochemistry(2nd ed.). New York: Worth.ISBN978-0-87901-500-8.[page needed]
- ^Abaza I, Gebauer F (March 2008)."Trading translation with RNA-binding proteins".RNA.14(3): 404–9.doi:10.1261/rna.848208.PMC2248257.PMID18212021.
- ^Mattick JS, Makunin IV (April 2006)."Non-coding RNA".Human Molecular Genetics.15 Spec No 1 (90001): R17-29.doi:10.1093/hmg/ddl046.PMID16651366.
- ^abHunt AG, Xu R, Addepalli B, Rao S, Forbes KP, Meeks LR, Xing D, Mo M, Zhao H, Bandyopadhyay A, Dampanaboina L, Marion A, Von Lanken C, Li QQ (May 2008)."Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling".BMC Genomics.9:220.doi:10.1186/1471-2164-9-220.PMC2391170.PMID18479511.
- ^abDávila López M, Samuelsson T (January 2008)."Early evolution of histone mRNA 3′ end processing".RNA.14(1): 1–10.doi:10.1261/rna.782308.PMC2151031.PMID17998288.
- ^Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ (November 2002). "The human and mouse replication-dependent histone genes".Genomics.80(5): 487–98.doi:10.1016/S0888-7543(02)96850-3.PMID12408966.
- ^Saini HK, Griffiths-Jones S, Enright AJ (November 2007)."Genomic analysis of human microRNA transcripts".Proceedings of the National Academy of Sciences of the United States of America.104(45): 17719–24.Bibcode:2007PNAS..10417719S.doi:10.1073/pnas.0703890104.PMC2077053.PMID17965236.
- ^Yoshikawa M, Peragine A, Park MY, Poethig RS (September 2005)."A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis".Genes & Development.19(18): 2164–75.doi:10.1101/gad.1352605.PMC1221887.PMID16131612.
- ^Amaral PP, Mattick JS (August 2008). "Noncoding RNA in development".Mammalian Genome.19(7–8): 454–92.doi:10.1007/s00335-008-9136-7.PMID18839252.S2CID206956408.
- ^abcdBienroth S, Keller W, Wahle E (February 1993)."Assembly of a processive messenger RNA polyadenylation complex".The EMBO Journal.12(2): 585–94.doi:10.1002/j.1460-2075.1993.tb05690.x.PMC413241.PMID8440247.
- ^abLiu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH (2006)."Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis".Nucleic Acids Research.35(1): 234–46.doi:10.1093/nar/gkl919.PMC1802579.PMID17158511.
- ^Lutz CS (October 2008). "Alternative polyadenylation: a twist on mRNA 3′ end formation".ACS Chemical Biology.3(10): 609–17.doi:10.1021/cb800138w.PMID18817380.
- ^abBeaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D (July 2000)."Patterns of variant polyadenylation signal usage in human genes".Genome Research.10(7): 1001–10.doi:10.1101/gr.10.7.1001.PMC310884.PMID10899149.
- ^Brown KM, Gilmartin GM (December 2003)."A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im".Molecular Cell.12(6): 1467–76.doi:10.1016/S1097-2765(03)00453-2.PMID14690600.
- ^Yang Q, Gilmartin GM, Doublié S (June 2010)."Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing".Proceedings of the National Academy of Sciences of the United States of America.107(22): 10062–7.Bibcode:2010PNAS..10710062Y.doi:10.1073/pnas.1000848107.PMC2890493.PMID20479262.
- ^Yang Q, Coseno M, Gilmartin GM, Doublié S (March 2011)."Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping".Structure.19(3): 368–77.doi:10.1016/j.str.2010.12.021.PMC3056899.PMID21295486.
- ^Venkataraman K, Brown KM, Gilmartin GM (June 2005)."Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition".Genes & Development.19(11): 1315–27.doi:10.1101/gad.1298605.PMC1142555.PMID15937220.
- ^abMillevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S (October 2006)."An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries".The EMBO Journal.25(20): 4854–64.doi:10.1038/sj.emboj.7601331.PMC1618107.PMID17024186.
- ^abcShen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ (May 2008)."Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation".Nucleic Acids Research.36(9): 3150–61.doi:10.1093/nar/gkn158.PMC2396415.PMID18411206.
- ^Glover-Cutter K, Kim S, Espinosa J, Bentley DL (January 2008)."RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes".Nature Structural & Molecular Biology.15(1): 71–8.doi:10.1038/nsmb1352.PMC2836588.PMID18157150.
- ^Molecular Biology of the Cell, Chapter 6, "From DNA to RNA". 4th edition. Alberts B, Johnson A, Lewis J, et al. New York: Garland Science; 2002.
- ^Stumpf G, Domdey H (November 1996). "Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor".Science.274(5292): 1517–20.Bibcode:1996Sci...274.1517S.doi:10.1126/science.274.5292.1517.PMID8929410.S2CID34840144.
- ^Iseli C, Stevenson BJ, de Souza SJ, Samaia HB, Camargo AA, Buetow KH, Strausberg RL, Simpson AJ, Bucher P, Jongeneel CV (July 2002)."Long-range heterogeneity at the 3′ ends of human mRNAs".Genome Research.12(7): 1068–74.doi:10.1101/gr.62002.PMC186619.PMID12097343.
- ^Balbo PB, Bohm A (September 2007)."Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis".Structure.15(9): 1117–31.doi:10.1016/j.str.2007.07.010.PMC2032019.PMID17850751.
- ^Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L (April 2008)."Molecular dissection of mRNA poly(A) tail length control in yeast".Nucleic Acids Research.36(7): 2418–33.doi:10.1093/nar/gkn080.PMC2367721.PMID18304944.
- ^Wahle E (February 1995)."Poly(A) tail length control is caused by termination of processive synthesis".The Journal of Biological Chemistry.270(6): 2800–8.doi:10.1074/jbc.270.6.2800.PMID7852352.
- ^Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W (August 2002)."Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination".The EMBO Journal.21(15): 4125–35.doi:10.1093/emboj/cdf390.PMC126137.PMID12145212.
- ^Nag A, Narsinh K, Martinson HG (July 2007). "The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase".Nature Structural & Molecular Biology.14(7): 662–9.doi:10.1038/nsmb1253.PMID17572685.S2CID5777074.
- ^Tefferi A, Wieben ED, Dewald GW, Whiteman DA, Bernard ME, Spelsberg TC (August 2002). "Primer on medical genomics part II: Background principles and methods in molecular genetics".Mayo Clinic Proceedings.77(8): 785–808.doi:10.4065/77.8.785.PMID12173714.S2CID2237085.
- ^Coller JM, Gray NK, Wickens MP (October 1998)."mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation".Genes & Development.12(20): 3226–35.doi:10.1101/gad.12.20.3226.PMC317214.PMID9784497.
- ^abSiddiqui N, Mangus DA, Chang TC, Palermino JM, Shyu AB, Gehring K (August 2007)."Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein".The Journal of Biological Chemistry.282(34): 25067–75.doi:10.1074/jbc.M701256200.PMID17595167.
- ^Vinciguerra P, Stutz F (June 2004). "mRNA export: an assembly line from genes to nuclear pores".Current Opinion in Cell Biology.16(3): 285–92.doi:10.1016/j.ceb.2004.03.013.PMID15145353.
- ^Gray NK, Coller JM, Dickson KS, Wickens M (September 2000)."Multiple portions of poly(A)-binding protein stimulate translation in vivo".The EMBO Journal.19(17): 4723–33.doi:10.1093/emboj/19.17.4723.PMC302064.PMID10970864.
- ^Meaux S, Van Hoof A (July 2006)."Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay".RNA.12(7): 1323–37.doi:10.1261/rna.46306.PMC1484436.PMID16714281.
- ^Kargapolova Y, Levin M, Lackner K, Danckwardt S (June 2017)."sCLIP-an integrated platform to study RNA-protein interactomes in biomedical research: identification of CSTF2tau in alternative processing of small nuclear RNAs".Nucleic Acids Research.45(10): 6074–6086.doi:10.1093/nar/gkx152.PMC5449641.PMID28334977.
- ^abMeijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH (2007)."A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells".Nucleic Acids Research.35(19): e132.doi:10.1093/nar/gkm830.PMC2095794.PMID17933768.
- ^Lehner B, Sanderson CM (July 2004)."A protein interaction framework for human mRNA degradation".Genome Research.14(7): 1315–23.doi:10.1101/gr.2122004.PMC442147.PMID15231747.
- ^Wu L, Fan J, Belasco JG (March 2006)."MicroRNAs direct rapid deadenylation of mRNA".Proceedings of the National Academy of Sciences of the United States of America.103(11): 4034–9.Bibcode:2006PNAS..103.4034W.doi:10.1073/pnas.0510928103.PMC1449641.PMID16495412.
- ^Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF (April 2008)."Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation".Genetics.178(4): 2017–29.doi:10.1534/genetics.107.084558.PMC2323793.PMID18430932.
- ^Torabi, Seyed-Fakhreddin; Vaidya, Anand T.; Tycowski, Kazimierz T.; DeGregorio, Suzanne J.; Wang, Jimin; Shu, Mei-Di; Steitz, Thomas A.; Steitz, Joan A. (2021-02-05)."RNA stabilization by a poly(A) tail 3′-end binding pocket and other modes of poly(A)-RNA interaction".Science.371(6529): eabe6523.doi:10.1126/science.abe6523.ISSN0036-8075.PMC9491362.PMID33414189.S2CID231195473.
- ^Wilusz CJ, Wormington M, Peltz SW (April 2001). "The cap-to-tail guide to mRNA turnover".Nature Reviews Molecular Cell Biology.2(4): 237–46.doi:10.1038/35067025.PMID11283721.S2CID9734550.
- ^Jung MY, Lorenz L, Richter JD (June 2006)."Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein".Molecular and Cellular Biology.26(11): 4277–87.doi:10.1128/MCB.02470-05.PMC1489097.PMID16705177.
- ^Sakurai T, Sato M, Kimura M (November 2005). "Diverse patterns of poly(A) tail elongation and shortening of murine maternal mRNAs from fully grown oocyte to 2-cell embryo stages".Biochemical and Biophysical Research Communications.336(4): 1181–9.doi:10.1016/j.bbrc.2005.08.250.PMID16169522.
- ^Taft RA (January 2008)."Virtues and limitations of the preimplantation mouse embryo as a model system".Theriogenology.69(1): 10–6.doi:10.1016/j.theriogenology.2007.09.032.PMC2239213.PMID18023855.
- ^Richter JD (June 2007). "CPEB: a life in translation".Trends in Biochemical Sciences.32(6): 279–85.doi:10.1016/j.tibs.2007.04.004.PMID17481902.
- ^Piqué M, López JM, Foissac S, Guigó R, Méndez R (February 2008)."A combinatorial code for CPE-mediated translational control".Cell.132(3): 434–48.doi:10.1016/j.cell.2007.12.038.PMID18267074.S2CID16092673.
- ^Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M (June 2008)."PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila".Development.135(11): 1969–79.doi:10.1242/dev.021444.PMC9154023.PMID18434412.
- ^Tian B, Hu J, Zhang H, Lutz CS (2005)."A large-scale analysis of mRNA polyadenylation of human and mouse genes".Nucleic Acids Research.33(1): 201–12.doi:10.1093/nar/gki158.PMC546146.PMID15647503.
- ^Danckwardt S, Hentze MW, Kulozik AE (February 2008)."3′ end mRNA processing: molecular mechanisms and implications for health and disease".The EMBO Journal.27(3): 482–98.doi:10.1038/sj.emboj.7601932.PMC2241648.PMID18256699.
- ^abTian, Bin; Manley, James L. (2017)."Alternative polyadenylation of mRNA precursors".Nature Reviews. Molecular Cell Biology.18(1): 18–30.doi:10.1038/nrm.2016.116.ISSN1471-0080.PMC5483950.PMID27677860.
- ^Zhang, Haibo; Lee, Ju Youn; Tian, Bin (2005)."Biased alternative polyadenylation in human tissues".Genome Biology.6(12): R100.doi:10.1186/gb-2005-6-12-r100.ISSN1474-760X.PMC1414089.PMID16356263.
- ^Smibert, Peter; Miura, Pedro; Westholm, Jakub O.; Shenker, Sol; May, Gemma; Duff, Michael O.; Zhang, Dayu; Eads, Brian D.; Carlson, Joe; Brown, James B.; Eisman, Robert C. (2012)."Global patterns of tissue-specific alternative polyadenylation in Drosophila".Cell Reports.1(3): 277–289.doi:10.1016/j.celrep.2012.01.001.ISSN2211-1247.PMC3368434.PMID22685694.
- ^Lee, Ju Youn; Ji, Zhe; Tian, Bin (2008)."Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3'-end of genes".Nucleic Acids Research.36(17): 5581–5590.doi:10.1093/nar/gkn540.ISSN1362-4962.PMC2553571.PMID18757892.
- ^Ogorodnikov A, Kargapolova Y, Danckwardt S (June 2016)."Processing and transcriptome expansion at the mRNA 3′ end in health and disease: finding the right end".Pflügers Archiv.468(6): 993–1012.doi:10.1007/s00424-016-1828-3.PMC4893057.PMID27220521.
- ^Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (June 2008)."Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites".Science.320(5883): 1643–7.Bibcode:2008Sci...320.1643S.doi:10.1126/science.1155390.PMC2587246.PMID18566288.
- ^Tili E, Michaille JJ, Calin GA (April 2008)."Expression and function of micro-RNAs in immune cells during normal or disease state".International Journal of Medical Sciences.5(2): 73–9.doi:10.7150/ijms.5.73.PMC2288788.PMID18392144.
- ^Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B (November 2008)."MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene".Nucleic Acids Research.36(19): 6318–32.doi:10.1093/nar/gkn624.PMC2577349.PMID18835850.
- ^Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D (June 1980). "Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends".Cell.20(2): 293–301.doi:10.1016/0092-8674(80)90615-7.PMID6771018.S2CID7448467.
- ^Tian B, Pan Z, Lee JY (February 2007)."Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing".Genome Research.17(2): 156–65.doi:10.1101/gr.5532707.PMC1781347.PMID17210931.
- ^abShell SA, Hesse C, Morris SM, Milcarek C (December 2005)."Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection".The Journal of Biological Chemistry.280(48): 39950–61.doi:10.1074/jbc.M508848200.PMID16207706.
- ^Ogorodnikov A, Levin M, Tattikota S, Tokalov S, Hoque M, Scherzinger D, Marini F, Poetsch A, Binder H, Macher-Göppinger S, Probst HC, Tian B, Schaefer M, Lackner KJ, Westermann F, Danckwardt S (December 2018)."Transcriptome 3′ end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma".Nature Communications.9(1): 5331.Bibcode:2018NatCo...9.5331O.doi:10.1038/s41467-018-07580-5.PMC6294251.PMID30552333.
- ^Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB (November 2008)."HITS-CLIP yields genome-wide insights into brain alternative RNA processing".Nature.456(7221): 464–9.Bibcode:2008Natur.456..464L.doi:10.1038/nature07488.PMC2597294.PMID18978773.
- ^Hall-Pogar T, Liang S, Hague LK, Lutz CS (July 2007)."Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR".RNA.13(7): 1103–15.doi:10.1261/rna.577707.PMC1894925.PMID17507659.
- ^Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE (June 2007)."Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals".The EMBO Journal.26(11): 2658–69.doi:10.1038/sj.emboj.7601699.PMC1888663.PMID17464285.
- ^Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Gröne HJ, Schirmacher P, Hentze MW, Kulozik AE (February 2011)."p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing".Molecular Cell.41(3): 298–310.doi:10.1016/j.molcel.2010.12.032.PMID21292162.
- ^Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ (May 2008)."Regulation of alternative polyadenylation by genomic imprinting".Genes & Development.22(9): 1141–6.doi:10.1101/gad.473408.PMC2335310.PMID18451104.
- ^Marini F, Scherzinger D, Danckwardt S (2021)."TREND-DB-a transcriptome-wide atlas of the dynamic landscape of alternative polyadenylation".Nucleic Acids Research.49(D1): D:243–D253.doi:10.1093/nar/gkaa722.PMC7778938.PMID32976578.
- ^Reinisch KM, Wolin SL (April 2007). "Emerging themes in non-coding RNA quality control".Current Opinion in Structural Biology.17(2): 209–14.doi:10.1016/j.sbi.2007.03.012.PMID17395456.
- ^Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E (June 2011)."The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex".Cell.145(6): 890–901.doi:10.1016/j.cell.2011.05.010.PMC3115544.PMID21663793.
- ^LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (June 2005)."RNA degradation by the exosome is promoted by a nuclear polyadenylation complex".Cell.121(5): 713–24.doi:10.1016/j.cell.2005.04.029.PMID15935758.S2CID14898055.
- ^abMartin G, Keller W (November 2007)."RNA-specific ribonucleotidyl transferases".RNA.13(11): 1834–49.doi:10.1261/rna.652807.PMC2040100.PMID17872511.
- ^Slomovic S, Laufer D, Geiger D, Schuster G (2006)."Polyadenylation of ribosomal RNA in human cells".Nucleic Acids Research.34(10): 2966–75.doi:10.1093/nar/gkl357.PMC1474067.PMID16738135.
- ^Régnier P, Arraiano CM (March 2000). "Degradation of mRNA in bacteria: emergence of ubiquitous features".BioEssays.22(3): 235–44.doi:10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2.PMID10684583.S2CID26109164.
- ^abcAnantharaman V, Koonin EV, Aravind L (April 2002)."Comparative genomics and evolution of proteins involved in RNA metabolism".Nucleic Acids Research.30(7): 1427–64.doi:10.1093/nar/30.7.1427.PMC101826.PMID11917006.
- ^abSlomovic S, Portnoy V, Liveanu V, Schuster G (2006). "RNA Polyadenylation in Prokaryotes and Organelles; Different Tails Tell Different Tales".Critical Reviews in Plant Sciences.25(1): 65–77.Bibcode:2006CRvPS..25...65S.doi:10.1080/07352680500391337.S2CID86607431.
- ^Chang, Jeong Ho; Tong, Liang (2012)."Mitochondrial poly(A) polymerase and polyadenylation".Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms.1819(9–10): 992–997.doi:10.1016/j.bbagrm.2011.10.012.ISSN0006-3002.PMC3307840.PMID22172994.
- ^Chang SA, Cozad M, Mackie GA, Jones GH (January 2008)."Kinetics of polynucleotide phosphorylase: comparison of enzymes from Streptomyces and Escherichia coli and effects of nucleoside diphosphates".Journal of Bacteriology.190(1): 98–106.doi:10.1128/JB.00327-07.PMC2223728.PMID17965156.
- ^Nagaike T, Suzuki T, Ueda T (April 2008). "Polyadenylation in mammalian mitochondria: insights from recent studies".Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms.1779(4): 266–9.doi:10.1016/j.bbagrm.2008.02.001.PMID18312863.
- ^Walter M, Kilian J, Kudla J (December 2002)."PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts".The EMBO Journal.21(24): 6905–14.doi:10.1093/emboj/cdf686.PMC139106.PMID12486011.
- ^Portnoy V, Schuster G (2006)."RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R".Nucleic Acids Research.34(20): 5923–31.doi:10.1093/nar/gkl763.PMC1635327.PMID17065466.
- ^Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G (September 2003)."Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins".The Plant Cell.15(9): 2003–19.doi:10.1105/tpc.013326.PMC181327.PMID12953107.
- ^Slomovic S, Portnoy V, Schuster G (2008). "Chapter 24 Detection and Characterization of Polyadenylated RNA in Eukarya, Bacteria, Archaea, and Organelles".RNA Turnover in Bacteria, Archaea and Organelles.Methods in Enzymology. Vol. 447. pp. 501–20.doi:10.1016/S0076-6879(08)02224-6.ISBN978-0-12-374377-0.PMID19161858.
- ^Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G (December 2005)."RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus".EMBO Reports.6(12): 1188–93.doi:10.1038/sj.embor.7400571.PMC1369208.PMID16282984.
- ^Portnoy V, Schuster G (June 2008)."Mycoplasma gallisepticum as the first analyzed bacterium in which RNA is not polyadenylated".FEMS Microbiology Letters.283(1): 97–103.doi:10.1111/j.1574-6968.2008.01157.x.PMID18399989.
- ^Evguenieva-Hackenberg E, Roppelt V, Finsterseifer P, Klug G (December 2008). "Rrp4 and Csl4 are needed for efficient degradation but not for polyadenylation of synthetic and natural RNA by the archaeal exosome".Biochemistry.47(50): 13158–68.doi:10.1021/bi8012214.PMID19053279.
- ^abSlomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G (April 2008). "Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases".Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms.1779(4): 247–55.doi:10.1016/j.bbagrm.2007.12.004.PMID18177749.
- ^Poon, Leo L. M.; Pritlove, David C.; Fodor, Ervin; Brownlee, George G. (1 April 1999)."Direct Evidence that the Poly(A) Tail of Influenza A Virus mRNA Is Synthesized by Reiterative Copying of a U Track in the Virion RNA Template".Journal of Virology.73(4): 3473–3476.doi:10.1128/JVI.73.4.3473-3476.1999.PMC104115.PMID10074205.
- ^Wu, Hung-Yi; Ke, Ting-Yung; Liao, Wei-Yu; Chang, Nai-Yun (2013)."Regulation of Coronaviral Poly(A) Tail Length during Infection".PLOS ONE.8(7): e70548.Bibcode:2013PLoSO...870548W.doi:10.1371/journal.pone.0070548.PMC3726627.PMID23923003.
- ^Neeleman, Lyda; Olsthoorn, René C. L.; Linthorst, Huub J. M.; Bol, John F. (4 December 2001)."Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA".Proceedings of the National Academy of Sciences.98(25): 14286–14291.Bibcode:2001PNAS...9814286N.doi:10.1073/pnas.251542798.PMC64674.PMID11717411.
- ^Chen, Jun-Hao; Zhang, Rui-Hua; Lin, Shao-Li; Li, Peng-Fei; Lan, Jing-Jing; Song, Sha-Sha; Gao, Ji-Ming; Wang, Yu; Xie, Zhi-Jing; Li, Fu-Chang; Jiang, Shi-Jin (2018)."The Functional Role of the 3′ Untranslated Region and Poly(A) Tail of Duck Hepatitis a Virus Type 1 in Viral Replication and Regulation of IRES-Mediated Translation".Frontiers in Microbiology.9:2250.doi:10.3389/fmicb.2018.02250.PMC6167517.PMID30319572.
- ^"Inhibition of host poly(A)-binding protein by virus ~ ViralZone".viralzone.expasy.org.
- ^Edmonds, Mary;Abrams, Richard (April 1960)."Polynucleotide Biosynthesis: Formation of a Sequence of Adenylate Units from Adenosine Triphosphate by an Enzyme from Thymus Nuclei".Journal of Biological Chemistry.235(4): 1142–1149.doi:10.1016/S0021-9258(18)69494-3.
- ^Colgan DF, Manley JL (November 1997)."Mechanism and regulation of mRNA polyadenylation".Genes & Development.11(21): 2755–66.doi:10.1101/gad.11.21.2755.PMID9353246.
- ^abEdmonds, M(2002).A history of poly A sequences: from formation to factors to function.Progress in Nucleic Acid Research and Molecular Biology. Vol. 71. pp. 285–389.doi:10.1016/S0079-6603(02)71046-5.ISBN978-0-12-540071-8.PMID12102557.
- ^Edmonds, M.;Vaughan, M. H.; Nakazato, H. (1 June 1971)."Polyadenylic Acid Sequences in the Heterogeneous Nuclear RNA and Rapidly-Labeled Polyribosomal RNA of HeLa Cells: Possible Evidence for a Precursor Relationship".Proceedings of the National Academy of Sciences.68(6): 1336–1340.Bibcode:1971PNAS...68.1336E.doi:10.1073/pnas.68.6.1336.PMC389184.PMID5288383.
Further reading
[edit]- Danckwardt S, Hentze MW, Kulozik AE (February 2008)."3′ end mRNA processing: molecular mechanisms and implications for health and disease".The EMBO Journal.27(3): 482–98.doi:10.1038/sj.emboj.7601932.PMC2241648.PMID18256699.
External links
[edit] Media related toPolyadenylationat Wikimedia Commons
Media related toPolyadenylationat Wikimedia Commons
