Ribose
| |||
| |||
| Names | |||
|---|---|---|---|
| IUPAC name
D-Ribose
| |||
| Systematic IUPAC name
(2R,3R,4S,5R)-5-(hydroxymethyl)oxolane-2,3,4-triol | |||
| Other names
d-Ribose
| |||
| Identifiers | |||
3D model (JSmol)
|
| ||
| ChEMBL | |||
| ChemSpider |
| ||
| DrugBank | |||
| EC Number |
| ||
PubChemCID
|
|||
| UNII | |||
| |||
| |||
| Properties[1][2] | |||
| C5H10O5 | |||
| Molar mass | 150.13 | ||
| Appearance | White solid | ||
| Melting point | 95 °C (203 °F; 368 K) | ||
| 100g/L (25 °C, 77 °F) | |||
Chiral rotation([α]D)
|
−21.5° (H2O) | ||
| Related compounds | |||
Relatedaldopentoses
|
Arabinose Xylose Lyxose | ||
Related compounds
|
Deoxyribose | ||
Except where otherwise noted, data are given for materials in theirstandard state(at 25 °C [77 °F], 100 kPa).
| |||

Riboseis asimple sugarandcarbohydratewithmolecular formulaC5H10O5and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form,d-ribose,is a component of theribonucleotidesfrom whichRNAis built, and so this compound is necessary forcoding,decoding,regulationandexpressionofgenes.It has astructural analog,deoxyribose,which is a similarly essential component ofDNA.l-riboseis an unnatural sugar that was first prepared byEmil FischerandOscar Pilotyin 1891.[3]It was not until 1909 thatPhoebus LeveneandWalter Jacobsrecognised thatd-ribosewas anatural product,theenantiomerof Fischer and Piloty's product, and an essential component ofnucleic acids.[4][5][6]Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar,arabinose,of which ribose is anepimerat the 2' carbon; both names also relate togum arabic,from which arabinose was first isolated and from which they preparedl-ribose.[6][7]
Right:Fischer projectionof theopen chainforms ofd- andl- ribose
Like most sugars, ribose exists as a mixture ofcyclic formsinequilibriumwith its linear form, and these readily interconvert especially inaqueous solution.[8]The name "ribose" is used in biochemistry and biology to refer to all of these forms, though more specific names for each are used when required. In its linear form, ribose can be recognised as thepentosesugar with all of itshydroxylfunctional groupson the same side in itsFischer projection.d-Ribosehas these hydroxyl groups on the right hand side and is associated with thesystematic name(2R,3R,4R)-2,3,4,5-tetrahydroxypentanal,[9]whilstl-ribosehas its hydroxyl groups appear on the left hand side in a Fischer projection. Cyclisation of ribose occurs viahemiacetalformation due to attack on thealdehydeby the C4' hydroxyl group to produce afuranoseform or by the C5' hydroxyl group to produce apyranoseform. In each case, there are two possible geometric outcomes, named as α- and β- and known asanomers,depending on thestereochemistryat the hemiacetal carbon atom (the "anomeric carbon" ). At room temperature, about 76% ofd-riboseis present in pyranose forms[8]: 228 (α:β = 1:2)[10]and 24% in the furanose forms[8]: 228 (α:β = 1:3),[10]with only about 0.1% of the linear form present.[11][12]
Theribonucleosidesadenosine,cytidine,guanosine,anduridineare allderivativesof β-d-ribofuranose.Metabolically-importantspecies that includephosphorylatedribose includeADP,ATP,coenzyme A,[8]: 228–229 andNADH.cAMPandcGMPserve as secondary messengers in some signaling pathways and are also ribose derivatives. The ribosemoietyappears in some pharmaceutical agents, including the antibioticsneomycinandparomomycin.[10]
Synthesis and sources[edit]
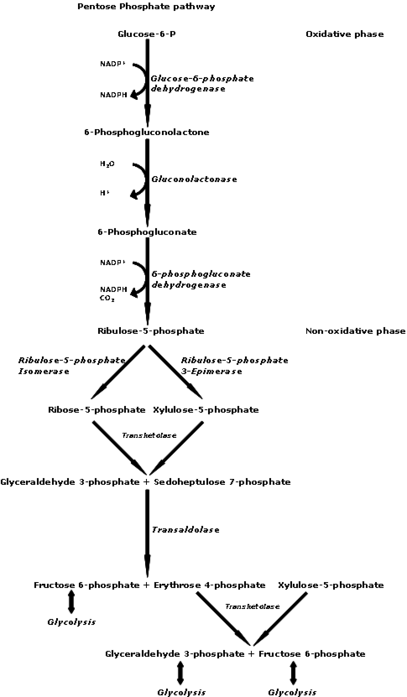
Ribose as its 5-phosphate ester is typically produced from glucose by thepentose phosphate pathway.In at least some archaea, alternative pathways have been identified.[13]
Ribose can be synthesized chemically, but commercial production relies on fermentation of glucose. Using genetically modified strains ofB. subtilis,90 g/liter of ribose can be produced from 200 g of glucose. The conversion entails the intermediacy of gluconate and ribulose.[14]
Ribose has been detected inmeteorites.[15][16]
Structure[edit]
Ribose is analdopentose(a monosaccharide containing fivecarbonatoms that, in itsopen chainform, has analdehydefunctional groupat one end). In the conventional numbering scheme for monosaccharides, the carbon atoms are numbered from C1' (in the aldehyde group) to C5'. Thedeoxyribosederivative found in DNA differs from ribose by having ahydrogenatom in place of thehydroxylgroup at C2'. This hydroxyl group performs a function inRNA splicing.
The "d- "in the named-ribose refers to thestereochemistryof thechiralcarbon atom farthest away from the aldehyde group (C4'). Ind-ribose, as in alld-sugars, this carbon atom has the same configuration as ind-glyceraldehyde.
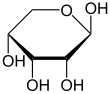
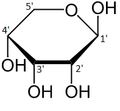
-
α-d-Ribopyranose
-
β-d-Ribopyranose
-
α-d-Ribofuranose
-
β-d-Ribofuranose
Relative abundance of forms of ribose in solution: β-d-ribopyranose (59%), α-d-ribopyranose (20%), β-d-ribofuranose (13%), α-d-ribofuranose (7%) and open chain (0.1%).[11]
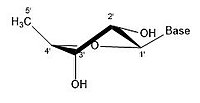
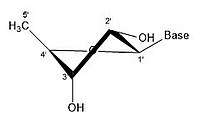
For ribose residues innucleosidesandnucleotide,the torsion angles for the rotation encompassing the bonds influence the configuration of the respective nucleoside and nucleotide. Thesecondary structureof a nucleic acid is determined by the rotation of its 7torsion angles.[17]Having a large amount of torsion angles allows for greater flexibility.
In closed ring riboses, the observed flexibility mentioned above is not observed because the ring cycle imposes a limit on the number of torsion angles possible in the structure.[17]Conformers of closed form riboses differ in regards to how the loneoxygenin the molecule is positioned respective to thenitrogenous base(also known as anucleobaseor just a base) attached to the ribose. If a carbon is facing towards the base, then the ribose is labeled as endo. If a carbon is facing away from the base, then the ribose is labeled as exo. If there is an oxygen molecule attached to the 2' carbon of a closed cycle ribose, then the exo confirmation is more stable because it decreases the interactions of the oxygen with the base.[17]The difference itself is quite small, but when looking at an entire chain of RNA the slight difference amounts to a sizable impact.
- Some pucker configurations of Ribose
-
2' endo
-
2' endo 3' exo
-
3' endo 2' exo
-
3' endo
A ribose molecule is typically represented as a planar molecule on paper. Despite this, it is typically non-planar in nature. Even between hydrogen atoms, the many constituents on a ribose molecule causesteric hindranceand strain between them. To relieve this crowding andring strain,the ring puckers, i.e. becomes non-planar.[18]This puckering is achieved by displacing an atom from the plane, relieving the strain and yielding a more stable configuration.[17]Puckering, otherwise known as the sugar ring conformation (specifically ribose sugar), can be described by the amplitude of pucker as well as thepseudorotationangle. The pseudo-rotation angle can be described as either "north (N)" or "south (S)" range. While both ranges are found in double helices, the north range is commonly associated with RNA and theA form of DNA.In contrast, the south range is associated withB form DNA.Z-DNAcontains sugars in both the north and south ranges.[19]When only a single atom is displaced, it is referred to as an "envelope" pucker. When two atoms are displaced, it is referred to as a "twist" pucker, in reference to the zigzag orientation.[20]In an "endo" pucker, the major displacement of atoms is on the β-face, the same side as the C4'-C5' bond and the base. In an "exo" pucker, the major displacement of atoms is on the α-face, on the opposite side of the ring. The major forms of ribose are the 3'-endo pucker (commonly adopted by RNA and A-form DNA) and 2'-endo pucker (commonly adopted by B-form DNA).[21]These ring puckers are developed from changes in ring torsion angles; there are infinite combinations of angles so therefore, there is an infinite number of transposable pucker conformations, each separated by disparate activation energies.
Functions[edit]
ATP is derived from ribose; it contains one ribose, threephosphategroups, and anadeninebase. ATP is created duringcellular respirationfromadenosine diphosphate(ATP with one less phosphate group).
Signaling pathways[edit]
Ribose is a building block in secondary signaling molecules such ascyclic adenosine monophosphate(cAMP) which is derived from ATP. One specific case in which cAMP is used is incAMP-dependent signaling pathways.In cAMP signaling pathways, either a stimulative or inhibitory hormone receptor is activated by asignal molecule.These receptors are linked to a stimulative or inhibitory regulativeG-protein.When a stimulative G-protein is activated,adenylyl cyclasecatalyzesATP into cAMP by using Mg2+or Mn2+.cAMP, a secondary messenger, then goes on to activateprotein kinase A,which is anenzymethat regulates cellmetabolism.Protein kinase A regulates metabolic enzymes byphosphorylationwhich causes a change in the cell depending on the original signal molecule. The opposite occurs when an inhibitory G-protein is activated; the G-protein inhibits adenylyl cyclase and ATP is not converted to cAMP.

Metabolism[edit]
Ribose is referred to as the "molecular currency" because of its involvement in intracellular energy transfers.[citation needed]For example,nicotinamide adenine dinucleotide(NAD),flavin adenine dinucleotide(FAD), andnicotinamide adenine dinucleotide phosphate(NADP) all contain thed-ribofuranosemoiety.They can each bederived fromd-ribose after it is converted tod-ribose 5-phosphateby the enzymeribokinase.[22][23]NAD, FAD, and NADP act as electron acceptors in biochemicalredoxreactions in major metabolic pathways includingglycolysis,thecitric acid cycle,fermentation,and theelectron transport chain.
Nucleotide biosynthesis[edit]
Nucleotides are synthesized through salvage orde novo synthesis.[24]Nucleotide salvageuses pieces of previously made nucleotides and re-synthesizes them for future use. In de novo, amino acids, carbon dioxide, folate derivatives, andphosphoribosyl pyrophosphate(PRPP) are used to synthesize nucleotides.[24]Both de novo and salvage require PRPP which is synthesized from ATP and ribose 5-phosphate by an enzyme calledPRPP synthetase.[24]
Modifications[edit]
Modifications in nature[edit]
Ribokinasecatalyzes the conversion ofd-ribose tod-ribose 5-phosphate.Once converted,d-ribose-5-phosphate is available for the manufacturing of theamino acidstryptophanandhistidine,or for use in thepentose phosphate pathway.The absorption ofd-ribose is 88–100% in the small intestines (up to 200 mg/kg·h).[25]
One important modification occurs at the C2' position of the ribose molecule. By adding anO-alkylgroup, the nuclear resistance of the RNA is increased because of additional stabilizing forces. These forces are stabilizing because of the increase ofintramolecular hydrogen bondingand an increase in theglycosidic bondstability.[26]The resulting increase of resistance leads to increases in thehalf-lifeofsiRNAand the potential therapeutic potential in cells and animals.[27]Themethylationof ribose at particular sites is correlated with a decrease in immune stimulation.[28]
Synthetic modifications[edit]
Along with phosphorylation, ribofuranose molecules can exchange their oxygen withseleniumandsulfurto produce similar sugars that only vary at the 4' position. These derivatives are morelipophilicthan the original molecule. Increased lipophilicity makes these species more suitable for use in techniques such asPCR,RNA aptamerpost-modification,antisense technology,and for phasingX-ray crystallographicdata.[27]
Similar to the 2' modifications in nature, a synthetic modification of ribose includes the addition offluorineat the 2' position. Thisfluorinatedribose acts similar to the methylated ribose because it is capable of suppressing immune stimulation depending on the location of the ribose in the DNA strand.[26]The big difference between methylation and fluorination, is the latter only occurs through synthetic modifications. The addition of fluorine leads to an increase in the stabilization of the glycosidic bond and an increase of intramolecular hydrogen bonds.[26]
Medical uses[edit]
d-ribose has been suggested for use in management ofcongestive heart failure[29](as well as other forms of heart disease) and forchronic fatigue syndrome(CFS), also called myalgic encephalomyelitis (ME) in an open-label non-blinded, non-randomized, and non-crossover subjective study.[30]
Supplementald-ribose can bypass part of thepentose phosphate pathway,an energy-producing pathway, to produced-ribose-5-phosphate. The enzymeglucose-6-phosphate-dehydrogenase(G-6-PDH) is often in short supply in cells, but more so in diseased tissue, such as inmyocardialcells in patients with cardiac disease. The supply ofd-ribose in themitochondriais directly correlated with ATP production; decreasedd-ribose supply reduces the amount of ATP being produced. Studies suggest that supplementingd-ribose following tissue ischemia (e.g. myocardial ischemia) increases myocardial ATP production, and therefore mitochondrial function. Essentially, administering supplementald-ribose bypasses an enzymatic step in the pentose phosphate pathway by providing an alternate source of 5-phospho-d-ribose 1-pyrophosphatefor ATP production. Supplementald-ribose enhances recovery of ATP levels while also reducing cellular injury in humans and other animals. One study suggested that the use of supplementald-ribose reduces the instance ofanginain men with diagnosedcoronary artery disease.[31]d-Ribose has been used to treat manypathologicalconditions, such as chronic fatigue syndrome,fibromyalgia,and myocardial dysfunction. It is also used to reduce symptoms of cramping, pain, stiffness, etc. after exercise and to improve athletic performance[citation needed].
References[edit]
- ^The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals(11th ed.), Merck, 1989,ISBN091191028X,8205
- ^Weast, Robert C., ed. (1981).CRC Handbook of Chemistry and Physics(62nd ed.). Boca Raton, FL: CRC Press. p. C-506.ISBN0-8493-0462-8.
- ^Fischer, Emil;Piloty, Oscar(1891)."Ueber eine neue Pentonsäure und die zweite inactive Trioxyglutarsäure"[About a new pentonic acid and the second inactive trioxyglutaric acid].Berichte der deutschen chemischen Gesellschaft(in German).24(2): 4214–4225.doi:10.1002/cber.189102402322.Archivedfrom the original on 4 June 2020.Retrieved12 March2020.
- ^Levene, P. A.;Jacobs, W. A.(1909). "Über Inosinsäure" [About inosic acid].Berichte der deutschen chemischen Gesellschaft(in German).42(1): 1198–1203.doi:10.1002/cber.190904201196.
- ^Levene, P. A.;Jacobs, W. A.(1909). "Über die Pentose in den Nucleinsäuren" [About the pentose in the nucleic acids].Berichte der deutschen chemischen Gesellschaft(in German).42(3): 3247–3251.doi:10.1002/cber.19090420351.
- ^abJeanloz, Roger W.;Fletcher, Hewitt G. (1951)."The Chemistry of Ribose".InHudson, Claude S.;Cantor, Sidney M. (eds.).Advances in Carbohydrate Chemistry.Vol. 6.Academic Press.pp. 135–174.doi:10.1016/S0096-5332(08)60066-1.ISBN9780080562650.PMID14894350.Archivedfrom the original on 26 October 2023.Retrieved15 December2019.
- ^Nechamkin, Howard (1958). "Some interesting etymological derivations of chemical terminology".Science Education.42(5): 463–474.Bibcode:1958SciEd..42..463N.doi:10.1002/sce.3730420523.
- ^abcdDewick, Paul M. (2013)."Oxygen as a Nucleophile: Hemicetals, Hemiketals, Acetals and Ketals".Essentials of Organic Chemistry: For Students of Pharmacy, Medicinal Chemistry and Biological Chemistry.John Wiley & Sons.pp. 224–234.ISBN9781118681961.Archivedfrom the original on 26 October 2023.Retrieved15 December2019.
- ^Leigh, Jeffery (July–August 2012)."Non-IUPAC Nomenclature Systems".Chemistry International.34(4).International Union of Pure and Applied Chemistry.Archivedfrom the original on 5 December 2019.Retrieved15 December2019.
- ^abcBhutani, S. P. (2019)."Aldopentoses—The Sugars of Nucleic Acids".Chemistry of Biomolecules(2nd ed.).CRC Press.pp. 63–65.ISBN9781000650907.Archivedfrom the original on 26 October 2023.Retrieved15 December2019.
- ^abDrew, Kenneth N.; Zajicek, Jaroslav; Bondo, Gail; Bose, Bidisha; Serianni, Anthony S. (February 1998)."13C-labeled aldopentoses: detection and quantitation of cyclic and acyclic forms by heteronuclear 1D and 2D NMR spectroscopy ".Carbohydrate Research.307(3–4): 199–209.doi:10.1016/S0008-6215(98)00040-8.
- ^de Wulf, P.; Vandamme, E. J. (1997). "Microbial Synthesis of ᴅ-Ribose: Metabolic Deregulation and Fermentation Process".Advances in Applied Microbiology.44:167–214.doi:10.1016/S0065-2164(08)70462-3.ISBN9780120026449.
- ^Tumbula, D. L.; Teng, Q.; Bartlett, M. G.; Whitman, W. B. (1997)."Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis".Journal of Bacteriology.179(19): 6010–6013.doi:10.1128/jb.179.19.6010-6013.1997.PMC179501.PMID9324245.
- ^Wulf, P. De; Vandamme, E. J. (1997). "Production of d -ribose by fermentation".Applied Microbiology and Biotechnology.48(2): 141–148.doi:10.1007/s002530051029.hdl:11572/262019.PMID9299771.S2CID34340369.
- ^Steigerwald, Bill; Jones, Nancy; Furukawa, Yoshihiro (18 November 2019)."First Detection of Sugars in Meteorites Gives Clues to Origin of Life".NASA.Archivedfrom the original on 15 January 2021.Retrieved18 November2019.
- ^Furukawa, Yoshihiro; Chikaraishi, Yoshito; Ohkouchi, Naohiko; Ogawa, Nanako O.; Glavin, Daniel P.; Dworkin, Jason P.; Abe, Chiaki; Nakamura, Tomoki (2019)."Extraterrestrial ribose and other sugars in primitive meteorites".Proceedings of the National Academy of Sciences of the United States of America.116(49): 24440–24445.Bibcode:2019PNAS..11624440F.doi:10.1073/pnas.1907169116.PMC6900709.PMID31740594.
- ^abcdBloomfield, Victor; Crothers, Donald; Tinoco, Ignacio (2000).Nucleic Acids: Structures, Properties, and Functions.University Science Books. pp.19–25.ISBN9780935702491.
- ^Voet, Donald; Voet, Judith (2011).Biochemistry.John Wiley & Sons, Inc. pp.1152,1153.ISBN978-0470570951.
- ^Foloppe, Nicolas; MacKerell, Alexander D. (August 1998). "Conformational Properties of the Deoxyribose and Ribose Moieties of Nucleic Acids: A Quantum Mechanical Study".The Journal of Physical Chemistry B.102(34): 6669–6678.doi:10.1021/jp9818683.ISSN1520-6106.
- ^"Nucleic acid architecture".fbio.uh.cu.Archived fromthe originalon 17 May 2018.Retrieved8 October2019.
- ^Neidle, Stephen (2008). "The Building-Blocks of DNA and RNA". In Neidle, Stephen (ed.).Principles of Nucleic Acid Structure.Academic Press.pp.20–37.doi:10.1016/B978-012369507-9.50003-0.ISBN9780123695079.
- ^Bork, Peer;Sander, Chris;Valencia, Alfonso(1993)."Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases".Protein Science.2(1): 31–40.doi:10.1002/pro.5560020104.PMC2142297.PMID8382990.
- ^Park, Jae; Gupta, Radhey S. (2008)."Adenosine kinase and ribokinase – the RK family of proteins".Cellular and Molecular Life Sciences.65(18): 2875–2896.doi:10.1007/s00018-008-8123-1.PMC11131688.PMID18560757.S2CID11439854.
- ^abcPuigserver, Pere (2018). "Signaling Transduction and Metabolomics". In Hoffman, Ronald; Benz, Edward J.; Silberstein, Leslie E.; Heslop, Helen E. (eds.).Hematology(7th ed.). Elsevier. pp. 68–78.doi:10.1016/B978-0-323-35762-3.00007-X.ISBN9780323357623.
- ^"Herbal Remedies, Supplements A-Z Index".PDRHealth.PDR, LLC. Archived fromthe originalon 11 October 2008.
- ^abcHamlow, Lucas; He, Chenchen; Fan, Lin; Wu, Ranran; Yang, Bo; Rodgers, M. T.; Berden, Giel; Oomens, J. (June 2015).Structual [sic] Effects of Cytidine 2'-Ribose Modifications as Determined by Irmpd Action Spectroscopy.70th International Symposium on Molecular Spectroscopy.University of Illinois Urbana-Champaign.Bibcode:2015isms.confEMI13H.doi:10.15278/isms.2015.MI13.
- ^abEvich, Marina; Spring-Connell, Alexander M.; Germann, Markus W. (27 January 2017)."Impact of modified ribose sugars on nucleic acid conformation and function".Heterocyclic Communications.23(3): 155–165.doi:10.1515/hc-2017-0056.ISSN2191-0197.S2CID91052034.
- ^Peacock, Hayden; Fucini, Raymond V.; Jayalath, Prasanna; Ibarra-Soza, José M.; Haringsma, Henry J.; Flanagan, W. Michael; Willingham, Aarron; Beal, Peter A. (2011)."Nucleobase and Ribose Modifications Control Immunostimulation by a MicroRNA-122-mimetic RNA".Journal of the American Chemical Society.133(24): 9200–9203.doi:10.1021/ja202492e.PMC3116021.PMID21612237.
- ^Omran, Heyder; McCarter, Dean; St Cyr, John; Lüderitz, Berndt (2004)."ᴅ-Ribose aids congestive heart failure patients".Experimental & Clinical Cardiology.Summer (9(2)): 117–118.PMC2716264.PMID19641697.
- ^Teitelbaum, Jacob E.; Johnson, Clarence; St Cyr, John (26 November 2006). "The use of ᴅ-ribose in chronic fatigue syndrome and fibromyalgia: a pilot study".The Journal of Alternative and Complementary Medicine.12(9): 857–862.CiteSeerX10.1.1.582.4800.doi:10.1089/acm.2006.12.857.PMID17109576.
- ^"Ribose".wa.kaiserpermanente.org.Archivedfrom the original on 3 March 2021.Retrieved7 October2019.