WPGMA
WPGMA(WeightedPairGroupMethod withArithmetic Mean) is a simple agglomerative (bottom-up)hierarchical clusteringmethod, generally attributed toSokalandMichener.[1]
The WPGMA method is similar to itsunweightedvariant, theUPGMAmethod.
Algorithm
[edit]The WPGMA algorithm constructs a rooted tree (dendrogram) that reflects the structure present in a pairwisedistance matrix(or asimilarity matrix). At each step, the nearest two clusters, sayand,are combined into a higher-level cluster.Then, its distance to another clusteris simply the arithmetic mean of the average distances between members ofandandand:
The WPGMA algorithm produces rooted dendrograms and requires a constant-rate assumption: it produces anultrametrictree in which the distances from the root to every branch tip are equal. Thisultrametricityassumption is called themolecular clockwhen the tips involveDNA,RNAandproteindata.
Working example
[edit]This working example is based on aJC69genetic distance matrix computed from the5S ribosomal RNAsequence alignment of five bacteria:Bacillus subtilis(),Bacillus stearothermophilus(),Lactobacillusviridescens(),Acholeplasmamodicum(), andMicrococcus luteus().[2][3]
First step
[edit]- First clustering
Let us assume that we have five elementsand the following matrixof pairwise distances between them:
| a | b | c | d | e | |
|---|---|---|---|---|---|
| a | 0 | 17 | 21 | 31 | 23 |
| b | 17 | 0 | 30 | 34 | 21 |
| c | 21 | 30 | 0 | 28 | 39 |
| d | 31 | 34 | 28 | 0 | 43 |
| e | 23 | 21 | 39 | 43 | 0 |
In this example,is the smallest value of,so we join elementsand.
- First branch length estimation
Letdenote the node to whichandare now connected. Settingensures that elementsandare equidistant from.This corresponds to the expectation of theultrametricityhypothesis. The branches joiningandtothen have lengths(see the final dendrogram)
- First distance matrix update
We then proceed to update the initial distance matrixinto a new distance matrix(see below), reduced in size by one row and one column because of the clustering ofwith. Bold values incorrespond to the new distances, calculated byaveraging distancesbetween each element of the first clusterand each of the remaining elements:
Italicized values inare not affected by the matrix update as they correspond to distances between elements not involved in the first cluster.
Second step
[edit]- Second clustering
We now reiterate the three previous steps, starting from the new distance matrix:
| (a,b) | c | d | e | |
|---|---|---|---|---|
| (a,b) | 0 | 25.5 | 32.5 | 22 |
| c | 25.5 | 0 | 28 | 39 |
| d | 32.5 | 28 | 0 | 43 |
| e | 22 | 39 | 43 | 0 |
Here,is the smallest value of,so we join clusterand element.
- Second branch length estimation
Letdenote the node to whichandare now connected. Because of the ultrametricity constraint, the branches joiningorto,andtoare equal and have the following length:
We deduce the missing branch length: (see the final dendrogram)
- Second distance matrix update
We then proceed to update thematrix into a new distance matrix(see below), reduced in size by one row and one column because of the clustering ofwith:
Of note, thisaverage calculationof the new distance does not account for the larger size of thecluster (two elements) with respect to(one element). Similarly:
The averaging procedure therefore gives differential weight to the initial distances of matrix.This is the reason why the method isweighted,not with respect to the mathematical procedure but with respect to the initial distances.
Third step
[edit]- Third clustering
We again reiterate the three previous steps, starting from the updated distance matrix.
| ((a,b),e) | c | d | |
|---|---|---|---|
| ((a,b),e) | 0 | 32.25 | 37.75 |
| c | 32.25 | 0 | 28 |
| d | 37.75 | 28 | 0 |
Here,is the smallest value of,so we join elementsand.
- Third branch length estimation
Letdenote the node to whichandare now connected. The branches joiningandtothen have lengths(see the final dendrogram)
- Third distance matrix update
There is a single entry to update:
Final step
[edit]The finalmatrix is:
| ((a,b),e) | (c,d) | |
|---|---|---|
| ((a,b),e) | 0 | 35 |
| (c,d) | 35 | 0 |
So we join clustersand.
Letdenote the (root) node to whichandare now connected. The branches joiningandtothen have lengths:
We deduce the two remaining branch lengths:
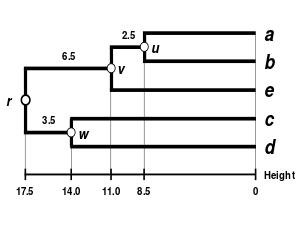
The WPGMA dendrogram
[edit]
The dendrogram is now complete. It is ultrametric because all tips (to) are equidistant from:
The dendrogram is therefore rooted by,its deepest node.
Comparison with other linkages
[edit]Alternative linkage schemes includesingle linkage clustering,complete linkage clustering,andUPGMA average linkage clustering.Implementing a different linkage is simply a matter of using a different formula to calculate inter-cluster distances during the distance matrix update steps of the above algorithm. Complete linkage clustering avoids a drawback of the alternative single linkage clustering method - the so-calledchaining phenomenon,where clusters formed via single linkage clustering may be forced together due to single elements being close to each other, even though many of the elements in each cluster may be very distant to each other. Complete linkage tends to find compact clusters of approximately equal diameters.[4]
 |

|
 |
 |
| Single-linkage clustering | Complete-linkage clustering | Average linkage clustering: WPGMA. | Average linkage clustering:UPGMA |
See also
[edit]- Neighbor-joining
- Molecular clock
- Cluster analysis
- Single-linkage clustering
- Complete-linkage clustering
- Hierarchical clustering
References
[edit]- ^Sokal,Michener(1958)."A statistical method for evaluating systematic relationships".University of Kansas Science Bulletin.38:1409–1438.
- ^Erdmann VA, Wolters J (1986)."Collection of published 5S, 5.8S and 4.5S ribosomal RNA sequences".Nucleic Acids Research.14 Suppl (Suppl): r1–59.doi:10.1093/nar/14.suppl.r1.PMC341310.PMID2422630.
- ^Olsen GJ (1988). "Phylogenetic analysis using ribosomal RNA".Ribosomes.Methods in Enzymology. Vol. 164. pp. 793–812.doi:10.1016/s0076-6879(88)64084-5.ISBN978-0-12-182065-7.PMID3241556.
- ^Everitt, B. S.; Landau, S.; Leese, M. (2001).Cluster Analysis. 4th Edition.London: Arnold. pp. 62–64.








































