YIF1A
| YIF1A | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | YIF1A,54TM, FinGER7, YIF1, YIF1P, Yip1 interacting factor homolog A, membrane trafficking protein | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM:611484;MGI:1915340;HomoloGene:56295;GeneCards:YIF1A;OMA:YIF1A - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Protein YIF1Ais aYip1 domain familyproteins that in humans is encoded by theYIF1Agene.[5][6][7][8]
Gene
[edit]YIF1A (Yip1 interacting factor homolog A) is also known as YIF1, YIF1P, FinGER7, and 54TM.[9]It has 4,591 base pairs with 8 exons, and it is located on the minus strand ofchromosome11, at 11q13.2, in humans.[10]
Promoters
[edit]There are four predictedpromoterfor YIIF1A.[11]The predicted promoter region with highest confidence is GXP_50494 and has 1252 base pairs long; it extends past the first exon of YIF1A. This promoter is located on the minus strand of chromosome 11.
Transcription factors
[edit]The promoter of YIF1A transcript variant 1 contains numerous transcription factor binding sites.[12]Transcription factors predicted to bind to the promoter region include the following.
- Acute myeloid leukemia 1 protein, RUNX1 (runt-related transcription factor 1)
- Zinc finger protein 263, ZKSCAN12 (zinc finger protein with KRAB and SCAN domains 12)
- E2F transcription factor 1
- EGR1, early growth response 1
- GATA-binding factor 1
- Transcription factor CP2-like 1 (LBP-9)
- X-box binding protein RFX1
- Estrogen response elements (ER Alpha ), IR3 sites
- Lactotransferrin and deltalactoferrin, growth-inhibiting protein 12
- TGFB-inducible early growth response protein 1 (KLF10)
Expression
[edit]The expression of YIF1A is highest in theduodenumandliver.It is also expressed at moderate levels in tissues including the colon, ovary, pancreases, spleen, and esophagus, and expressed at lower levels in a variety of other tissues.[13][14][15]NCBI GeoProfile data provide the tissue expression graph for YIF1A in humans; it also indicates that YIF1A is expressed at moderately to moderately low across all other tissues.[16]
mRNA
[edit]
YIF1A has isoforms 1 and 2, with exons 8 and 7 respectively.[10]The two transcripts undergo alternate splicing and are translated into proteins with 293 and 241 amino acids, respectively.[17][18]
RNA-binding proteins
[edit]The 5' untranslated region has predicted sites for binding by RBXM,EIF4B,andFUS.The 3' untranslated region has predicted sites for binding byELAVL1,which is AU rich elements and regulate mRNA stability.[19]
Protein
[edit]The longest proteinisoformof YIF1A is 293 amino acids in length. It has an observed molecular weight of approximately 32.0 kDa with a predictedisoelectric pointof approximately 8.98.[18][20][21]
Composition
[edit]YIF1 is a very normal protein in terms of the amino acid quantities it contains. The composition of each amino acid residue is similar to its average relative composition among human proteins. There are no charge clusters, runs, or patterns. There is a repetitive structure for protein YIF1A at [ 201- 204 and 288- 291 ] TFHL.[20]
Domain and motifs
[edit]YIF1A has a conserved domain, pfam03878 (AA 57 →287).[10]Within the domain, there are 5 transmembrane domains, 3 non-cytosolic domains, and 3 cytosolic domains. It has been hypothesized that there is a possible role in transport between theendoplasmic reticulumandGolgi.[9]
Structure
[edit]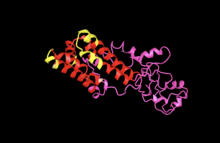
The structure of YIF1A consist of approximately 59% Alpha -helices, with TM helix and disordered regions making up the rest of the structure; no beta- strand was predicted.[24]
Localization
[edit]YIF1A's predicted location is in the endoplasmic reticulum, with intracellular N-terminus and an extracellular C-terminus.[25][26]
Post-translational modifications
[edit]YIF1A undergoes methionine cleavage and N-terminalacetylation,which is one of the most common post translation modifications of eukaryotic proteins.[27]It also phosphorylated by unspecified kinases at several sites.[28]Three glycation site is predicted in lysine residue(lys 104,161, and 211).[29]YIF1A undergoes O-ß-GlcNAc modification at 5 sites, 1 of them being Yin-Yang sites.[30]
Interacting protein
[edit]Based onfluorescence microscopy,validated two hybrid, and anti tag coimmunoprecipitation, the protein that is most likely to interact with YIF1A areGPR37,SEC23IP,REEP2,andYIPF5.Studies suggest that interaction between VAPB and YIF1A control membrane delivery into dendrites.[31]It also participates in ER unfolded protein response (UPR) by inducing ERN1/IRE1.[32]Additionally, the YIF1A protein interacts with the M protein ofSARS-Cov-2.[33]

Homology
[edit]YIF1A has a singleParalogcalled YIF1B, which is located on human chromosome 19.[9]YIF1A has 238 identifiedorthologs.[34]The ortholog contains vertebrates such as mammals,amphibians,andreptiles.It also hasinvertebratesspecies such as Insecta,Anthozoa,andAscidiacea.No ortholog was found in protists, bacteria, or archaea.
The following table provides a sample of the ortholog of YIF1A.
| Genus and species | Accession Number[10] | Date of Divergence (MYA)[35] | Sequence Length(AA) | Sequence Identity[36] |
|---|---|---|---|---|
| Homo sapiens(Human) | NP_065203 | 0 | 293 | 100 |
| Aotus nancymaae(Ma's night monkey) | XP_012318344 | 43 | 317 | 94 |
| Mus musculus(Mouse) | NP_080829 | 90 | 293 | 93 |
| Sus scrofa(Wild Boar) | XP_013849519 | 96 | 311 | 92 |
| Delphinapterus leucas(White whale) | XP_022447094 | 96 | 306 | 91 |
| Phascolarctos cinereus(Koala) | XP_020823757 | 159 | 293 | 88 |
| Ornithorhynchus anatinus(Platypus) | XP_028915982 | 177 | 293 | 88 |
| Chelonia mydas(Green turtle) | XP_007056281 | 312 | 240 | 78 |
| Chrysemys picta bellii(Painted turtle) | XP_005305497 | 312 | 293 | 73 |
| Microcaecilia unicolor(Amph.) | XP_029470520 | 352 | 306 | 72 |
| Rhinatrema bivittatum(Two-lined caecilian) | XP_029470520 | 352 | 307 | 71 |
| Latimeria chalumnae(Gombessa) | XP_014345204 | 413 | 296 | 71 |
| Salmo trutta(Brown trou) | XP_029585843 | 435 | 309 | 70 |
| Echeneis naucrates(live sharksucker) | XP_029368074 | 435 | 308 | 66 |
| Danio rerio(Zebrafish) | NP_956225 | 435 | 307 | 65 |
| Maylandia zebra(zebra mbuna) | XP_004545672 | 435 | 308 | 63 |
| Saccharomyces cerevisiaeS288C (Baker's yeast) | NP_014136 | 1017 | 314 | 33 |
| Physcomitrium patens(moss) | XP_024362517 | 1275 | 282 | 30 |
References
[edit]- ^abcGRCh38: Ensembl release 89: ENSG00000174851–Ensembl,May 2017
- ^abcGRCm38: Ensembl release 89: ENSMUSG00000024875–Ensembl,May 2017
- ^"Human PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^"Mouse PubMed Reference:".National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^Vitale G, Alexandrov K, Ullrich O, Horiuchi H, Giner A, Dobson C, et al. (Jan 1997). "The GDP/GTP cycle of Rab5 in the regulation of endocytotic membrane traffic".Cold Spring Harbor Symposia on Quantitative Biology.60:211–20.doi:10.1101/SQB.1995.060.01.024.PMID8824393.
- ^Matern H, Yang X, Andrulis E, Sternglanz R, Trepte HH, Gallwitz D (September 2000)."A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting".The EMBO Journal.19(17): 4485–92.doi:10.1093/emboj/19.17.4485.PMC302084.PMID10970842.
- ^Yoshida Y, Suzuki K, Yamamoto A, Sakai N, Bando M, Tanimoto K, et al. (November 2008). "YIPF5 and YIF1A recycle between the ER and the Golgi apparatus and are involved in the maintenance of the Golgi structure".Experimental Cell Research.314(19): 3427–43.doi:10.1016/j.yexcr.2008.07.023.PMID18718466.
- ^"Entrez Gene: YIF1A Yip1 interacting factor homolog A (S. cerevisiae)".
- ^abc"YIF1A related genes - GeneCards Search Results".genecards.org.Retrieved2020-06-21.
- ^abcd"YIF1A - Gene - NCBI".ncbi.nlm.nih.gov.Retrieved2020-06-21.
- ^"Genomatix: Genome Annotation and Browser: Query Input".genomatix.de.Retrieved2020-07-30.[permanent dead link]
- ^"Genomatix: MatInspector Input".genomatix.de.Retrieved2020-08-03.[permanent dead link]
- ^Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. (February 2014)."Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics".Molecular & Cellular Proteomics.13(2): 397–406.doi:10.1074/mcp.M113.035600.PMC3916642.PMID24309898.
- ^Duff MO, Olson S, Wei X, Garrett SC, Osman A, Bolisetty M, et al. (May 2015)."Genome-wide identification of zero nucleotide recursive splicing in Drosophila".Nature.521(7552): 376–9.Bibcode:2015Natur.521..376D.doi:10.1038/nature14475.PMC4529404.PMID25970244.
- ^Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, et al. (June 2015)."Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development".Genome Biology.16(1): 126.doi:10.1186/s13059-015-0690-5.PMC4506483.PMID26076956.
- ^"GDS596 / 202418_at".ncbi.nlm.nih.gov.Retrieved2020-08-02.
- ^"protein YIF1A isoform 2 [Homo sapiens] - Protein - NCBI".ncbi.nlm.nih.gov.Retrieved2020-07-28.
- ^ab"protein YIF1A isoform 1 [Homo sapiens] - Protein - NCBI".ncbi.nlm.nih.gov.Retrieved2020-07-28.
- ^"RBPDB: The database of RNA-binding specificities".rbpdb.ccbr.utoronto.ca.Retrieved2020-08-01.
- ^ab"SAPS < Sequence Statistics < EMBL-EBI".ebi.ac.uk.Retrieved2020-07-28.
- ^"ExPASy - Compute pI/Mw tool".web.expasy.org.Retrieved2020-07-28.
- ^"I-TASSER server for protein structure and function prediction".zhanglab.ccmb.med.umich.edu.Retrieved2020-08-01.
- ^"iCn3D: Web-based 3D Structure Viewer".ncbi.nlm.nih.gov.Retrieved2020-08-01.
- ^"NPS@: GOR4 secondary structure prediction".npsa-prabi.ibcp.fr.Retrieved2020-07-28.
- ^"PredictProtein - Protein Sequence Analysis, Prediction of Structural and Functional Features".predictprotein.org.Retrieved2020-07-28.
- ^"Phobius".phobius.sbc.su.se.Retrieved2020-07-28.
- ^"TERMINUS - Welcome to terminus".terminus.unige.ch.Retrieved2020-07-28.
- ^"NetPhosK 1.0 Server".cbs.dtu.dk.Archived fromthe originalon 2021-07-09.Retrieved2020-07-28.
- ^"NetGlycate 1.0 Server - prediction results".cbs.dtu.dk.Retrieved2020-08-01.
- ^"YinOYang 1.2 Server".cbs.dtu.dk.Retrieved2020-07-28.
- ^Kuijpers M, Yu KL, Teuling E, Akhmanova A, Jaarsma D,Hoogenraad CC(July 2013)."The ALS8 protein VAPB interacts with the ER-Golgi recycling protein YIF1A and regulates membrane delivery into dendrites".The EMBO Journal.32(14): 2056–72.doi:10.1038/emboj.2013.131.PMC3715857.PMID23736259.
- ^"YIF1A protein (human) - STRING interaction network".string-db.org.Retrieved2020-07-29.
- ^Mahen R (2020-04-09)."A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing".doi:10.1242/prelights.18355.S2CID243418486.Retrieved2020-08-05.
{{cite journal}}:Cite journal requires|journal=(help) - ^"Nucleotide BLAST: Search nucleotide databases using a nucleotide query".blast.ncbi.nlm.nih.gov.Retrieved2020-08-03.
- ^"TimeTree:: The Timescale of Life".timetree.org.Retrieved2020-07-02.
- ^"Human BLAT Search".genome.ucsc.edu.Retrieved2020-07-02.
Further reading
[edit]- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides".Gene.138(1–2): 171–4.doi:10.1016/0378-1119(94)90802-8.PMID8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library".Gene.200(1–2): 149–56.doi:10.1016/S0378-1119(97)00411-3.PMID9373149.
- Gisler SM, Stagljar I, Traebert M, Bacic D, Biber J, Murer H (March 2001)."Interaction of the type IIa Na/Pi cotransporter with PDZ proteins".The Journal of Biological Chemistry.276(12): 9206–13.doi:10.1074/jbc.M008745200.PMID11099500.
- Calero M, Winand NJ, Collins RN (March 2002)."Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors".FEBS Letters.515(1–3): 89–98.doi:10.1016/S0014-5793(02)02442-0.PMID11943201.S2CID34319925.
- Breuza L, Halbeisen R, Jenö P, Otte S, Barlowe C, Hong W, Hauri HP (November 2004)."Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46".The Journal of Biological Chemistry.279(45): 47242–53.doi:10.1074/jbc.M406644200.PMID15308636.
- Jin C, Zhang Y, Zhu H, Ahmed K, Fu C, Yao X (August 2005). "Human Yip1A specifies the localization of Yif1 to the Golgi apparatus".Biochemical and Biophysical Research Communications.334(1): 16–22.doi:10.1016/j.bbrc.2005.06.051.PMID15990086.





