Archaea
| Archaea | |
|---|---|
| |
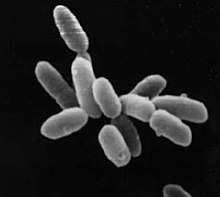
| Halobacteriumsp. strain NRC-1, each cell about 5μmlong | |
| Scientific classification | |
| Domain: | Archaea Woese,Kandler&Wheelis,1990 |
| Kingdoms[3]and phyla[4] | |
| |
| Synonyms | |
| |

TheArchaea(orArchea) are a group ofsingle-celledorganisms.The name comes from Greek αρχαία, "old ones". They are a major division of livingorganisms.
Archaea are tiny, simpleorganisms.They were originally discovered in extreme environments (extremophiles), but are now thought to be common to more average conditions. Many can survive at very high (over 80 °f) or very low temperatures, or highlysalty,acidicoralkalinewater. Some have been found ingeysers,black smokers,oil wells,and hot vents in the deep ocean. Recent research has foundammonia-eating archaea insoiland seawater.
In the past they had been classed withbacteriaasprokaryotes(orKingdom Monera) and namedarchaebacteria,but this is a mistake.[5]The Archaea have an independentevolutionary historyand show many differences in theirbiochemistryfrom other forms of life. They are now classified as a separate domain in thethree-domain system.In this system, the three distinct branches ofevolutionarydescent are the Archaea,BacteriaandEukaryota.
Archaea are, like bacteria,prokaryotes:single-celled organisms that do not havenucleiand cell organelles of theeukaryotetype.
Comparison to other domains
[change|change source]The following table compares some major characteristics of the three domains, to illustrate their similarities and differences.[6]Many of these characteristics are also discussed below.
| Property | Archaea | Bacteria | Eukarya |
|---|---|---|---|
| Cell membrane | Ether-linkedlipids,pseudopeptidoglycan | Ester-linked lipids,peptidoglycan | Ester-linked lipids, various structures |
| Genestructure | Circularchromosomes,similartranslationandtranscriptionto Eukarya | Circular chromosomes, unique translation and transcription | Multiple, linear chromosomes, similar translation and transcription to Archaea |
| Internalcellstructure | No membrane-boundorganelles(but questioned:[7]) ornucleus | No membrane-bound organelles or nucleus | Membrane-bound organelles and nucleus |
| Metabolism[8] | Various, withmethanogenesisunique to Archaea | Various, includingphotosynthesis,aerobic andanaerobic respiration,fermentation,andautotrophy | Photosynthesis, cellular respiration and fermentation |
| Reproduction | Asexual reproduction,horizontal gene transfer | Asexual reproduction, horizontal gene transfer | Sexualand asexual reproduction |
= Interesting facts about archaea:[9][10][11]
- No archaean species can dophotosynthesis.[12]
- Archaea only reproduceasexually.[13]
- Archaea show high levels ofhorizontal gene transferbetween lineages.[14][15]
- Many archaea live inextreme environments.
- Unlikebacteria,no archaea producespores.
- Archaea are common in theocean,and especially in theplankton.They make up to 20% of allmicrobialcells in the ocean.[16]p475
- Carl Woesediscovered the Archaea in 1978.
Related pages
[change|change source]Further reading
[change|change source]- Barry E.R. & Bell S.D. 2006. DNA replication in the Archaea.Microbiology and molecular biology reviews(MMBR) '70,876-887.
- Kelman L.M. & Kelman Z. 2003. Archaea: An archetype for replication initiation studies?Molecular microbiology,48,605-615.
Other websites
[change|change source]- Archaea-Citizendium
References
[change|change source]- ↑1.01.1"Taxa above the rank of class".List of Prokaryotic names with Standing in Nomenclature.Retrieved8 August2017.
- ↑Cavalier-Smith, T. (2014)."The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life".Cold Spring Harb. Perspect. Biol.6(9): a016006.doi:10.1101/cshperspect.a016006.PMC4142966.PMID25183828.
- ↑Petitjean C, Deschamps P, López-García P, Moreira D (December 2014)."Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota".Genome Biology and Evolution.7(1): 191–204.doi:10.1093/gbe/evu274.PMC4316627.PMID25527841.
- ↑"NCBI taxonomy page on Archaea".
{{cite journal}}:Cite journal requires|journal=(help) - ↑Pace NR (May 2006). "Time for a change".Nature.441(7091): 289.Bibcode:2006Natur.441..289P.doi:10.1038/441289a.PMID16710401.S2CID4431143.
- ↑Information is from Willey JM, Sherwood LM, Woolverton CJ.Microbiology7th ed. (2008), Ch. 19 pp. 474–475, except where noted.
- ↑Thomas Heimerl; et al. (13 June 2017)."A Complex endomembrane system in the ArchaeonIgnicoccus hospitalistapped byNanoarchaeum equitans".Frontiers in Microbiology.8:1072.doi:10.3389/fmicb.2017.01072.PMC5468417.PMID28659892.
- ↑Jurtshuk, Peter (1996). "Bacterial Metabolism".Medical Microbiology(4th ed.). Galveston (TX): University of Texas Medical Branch at Galveston.ISBN9780963117212.Retrieved5 November2014.
- ↑Howland, John L. (2000).The surprising Archaea: discovering another domain of life.Oxford: Oxford University Press.ISBN0-19-511183-4.
- ↑Garrett RA, Klenk H (2005).Archaea: evolution, physiology and molecular biology.WileyBlackwell.ISBN1-4051-4404-1.
- ↑Schaechter, M (2009).Archaea (overview) in The desk encyclopedia of microbiology, 2nd edition.San Diego and London: Elsevier Academic Press.ISBN978-0-12-374980-2.
- ↑Schäfer G.et al1999. Bioenergetics of the Archaea.Microbiol. Mol. Biol. Rev.63(3): 570–620.PMID 10477309.
- ↑de Queiroz K (2005)."Ernst Mayr and the modern concept of species".Proc. Natl. Acad. Sci. U.S.A.102(Suppl 1): 6600–7.Bibcode:2005PNAS..102.6600D.doi:10.1073/pnas.0502030102.PMC1131873.PMID15851674.
- ↑Eppley JM, Tyson GW, Getz WM, Banfield JF (2007)."Genetic exchange across a species boundary in the archaeal genus ferroplasma".Genetics.177(1): 407–16.doi:10.1534/genetics.107.072892.PMC2013692.PMID17603112.
{{cite journal}}:CS1 maint: multiple names: authors list (link) - ↑Papke RT, Zhaxybayeva O, Feil EJ, Sommerfeld K, Muise D, Doolittle WF (2007)."Searching for species in haloarchaea".Proc. Natl. Acad. Sci. U.S.A.104(35): 14092–7.Bibcode:2007PNAS..10414092P.doi:10.1073/pnas.0706358104.PMC1955782.PMID17715057.
{{cite journal}}:CS1 maint: multiple names: authors list (link) - ↑DeLong EF, Pace NR (2001). "Environmental diversity of bacteria and archaea".Syst. Biol.50(4): 470–8.doi:10.1080/106351501750435040.PMID12116647.Full text:[1]
