Abstract
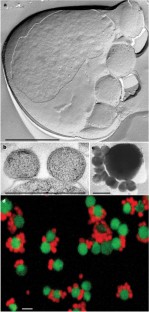
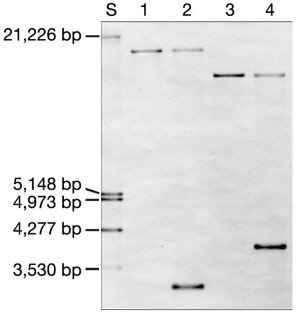
According to small subunit ribosomal RNA (ss rRNA) sequence comparisons all known Archaea belong to the phyla Crenarchaeota, Euryarchaeota, and—indicated only by environmental DNA sequences—to the ‘Korarchaeota’1,2.Here we report the cultivation of a new nanosized hyperthermophilic archaeon from a submarine hot vent. This archaeon cannot be attached to one of these groups and therefore must represent an unknown phylum which we name ‘Nanoarchaeota’ and species, which we name ‘Nanoarchaeum equitans’. Cells of ‘N. equitans’ are spherical, and only about 400 nm in diameter. They grow attached to the surface of a specific archaeal host, a new member of the genusIgnicoccus3.The distribution of the ‘Nanoarchaeota’ is so far unknown. Owing to their unusual ss rRNA sequence, members remained undetectable by commonly used ecological studies based on the polymerase chain reaction4.‘N. equitans’ harbours the smallest archaeal genome; it is only 0.5 megabases in size. This organism will provide insight into the evolution of thermophily, of tiny genomes and of interspecies communication.
This is a preview of subscription content,access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
185,98 € per year
only 3,65 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: Proposal for the domainsArchaea,BacteriaandEucarya.Proc. Natl Acad. Sci. USA87,4576–4579 (1990).
Barns, S. M., Delwiche, C. F., Palmers, J. D. & Pace, N. R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences.Proc. Natl Acad. Sci. USA93,9188–9193 (1996).
Huber, H. et al.Ignicoccusgen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species,Ignicoccus islandicussp. nov. andIgnicoccus pacificussp. nov.Int. J. Syst. Evol. Microbiol.50,2093–2100 (2000).
Barns, S. M., Fundyga, R. E., Jeffries, M. W. & Pace, N. R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment.Proc. Natl Acad. Sci. USA91,1609–1613 (1994).
Stetter, K. O. Microbial life in hyperthermal environments.Am. Soc. Microbiol. News61,285–290 (1995).
Stetter, K. O.Size Limits of Very Small Microorganisms; Proceedings of a Workshop(ed. Space studies board) 68–73 (National Academic Press, Washington DC, 1998).
Fricke, H., Giere, O., Stetter, K. O., Alfredsson, G. A. & Kristjansson, J. K. Hydrothermal vent communities at the shallow subpolar mid-Atlantic ridge.Mar. Biol.102,425–429 (1989).
Huber, H., Huber, G. & Stetter, K. O. A modified 4′,6′-diamidino-2-phenylindole fluorescence staining procedure suitable for the visualization of lithotrophic bacteria.Syst. Appl. Microbiol.6,105–106 (1985).
Ashkin, A., Dziedzic, J. M. & Yamane, T. Optical trapping and manipulation of single cells using infrared laser beams.Nature330,769–771 (1987).
Huber, R. et al. Isolation of a hyperthermophilic archaeum predicted byin situRNA analysis.Nature376,57–58 (1995).
Woese, C. R. Bacterial evolution.Microbiol. Rev.51,221–271 (1987).
Eder, W., Ludwig, W. & Huber, R. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea.Arch. Microbiol.172,213–218 (1999).
Sambrook, J. Molecular cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, New York, 1989).
Ludwig, W. et al. Bacterial phylogeny based on comparative sequence analysis.Electrophoresis19,554–568 (1998).
Burggraf, S. et al. Identifying members of the domainArchaeawith rRNA-targeted oligonucleotide probes.Appl. Environ. Microbiol.60,3112–3119 (1994).
Stahl, D. A. & Amann, R.Nucleic acid techniques in bacterial systematics(eds Stackebrandt, E. & Goodfellow, M.) 205–248 (Wiley, Chichester, 1991).
Preston, C. M., Wu, K. Y., Molinski, T. F. & DeLong, E. F. A psychrophilic crenarchaeon inhabits a marine sponge:Cenarchaeum symbiosumgen. nov., sp. nov.Proc. Natl Acad. Sci. USA93,6241–6246 (1996).
Boetius, A. et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane.Nature407,623–626 (2000).
Fraser, C. M. et al. the minimal gene complement ofMycoplasma genitalium.Science270,397–403 (1995).
Rohozinski, J., Girton, L. E. & Van Etten, J. L. Chlorella viruses contain linear nonpermutated double-stranded DNA genomes with covalently closed hairpin ends.Virology168,363–369 (1989).
Murphy, F. A. et al. (eds)Virus Taxonomy: Sixth Report of the International Committee on Taxonomy of Viruses(Springer, Vienna/New York, 1995).
Hutchinson, C. A. III et al. Global transposon mutagenesis and a minimalMycoplasmagenome.Science286,2165–2169 (1999).
Ochman, H. & Moran, N. A. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis.Science292,1096–1099 (2001).
Brosius, J., Dull, T J. Sleeter, D. D. & Noller, H. F. Gene organization and primary structure of a ribosomal RNA operon fromEscherichia coli.J. Mol. Biol.148,107–127 (1981).
Baumann, C., Judex, M., Huber, H. & Wirth, R. Estimation of genome sizes of hyperthermophiles.Extremophiles2,101–108 (1998).
Burggraf, S., Huber, H. & Stetter, K. O. Reclassification of the crenarchaeal orders and families in accordance with 16S ribosomal RNA sequence data.Int. J. Syst. Bacteriol.47,657–660 (1997).
Ludwig, W. & Strunk, O. ARB: A software environment for sequence data (2002); available athttp:// arb-home.de.
De Rijk, P. & De Wachter, R. RnaViz, a program for the visualisation of RNA secondary structure.Nucleic Acids Res.25,4679–4684 (1997).
Burggraf, S., Heyder, P. & Eis, N. A pivotal archaea group.Nature383,780 (1997).
Acknowledgements
We thank W. Ludwig and D. Prangishvili for stimulating discussions, S. Diller, S. Leptihn, M. Brandl, I. Wyschkony and P. Hummel for technical support, and B. Hedlund for critically reading the manuscript. We are grateful to the cruise leader P. Stoffers, the crew of RVPoseidonand the submersibleJagoteam for support during sampling, and the Icelandic government for a research permit. This study was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests
Rights and permissions
About this article
Cite this article
Huber, H., Hohn, M., Rachel, R.et al.A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature417,63–67 (2002). https://doi.org/10.1038/417063a
Received:
Accepted:
Issue Date:
DOI:https://doi.org/10.1038/417063a
This article is cited by
-
Uncovering the hidden world of nanosized archaea
Nature Reviews Microbiology(2023)
-
The cell biology of archaea
Nature Microbiology(2022)
-
The importance of biofilm formation for cultivation of a Micrarchaeon and its interactions with its Thermoplasmatales host
Nature Communications(2022)
-
Microbial diversity in extreme environments
Nature Reviews Microbiology(2022)
-
Culexarchaeia, a novel archaeal class of anaerobic generalists inhabiting geothermal environments
ISME Communications(2022)